The Journal of Advances in Parasitology
Research Article
Optimization of Polymerase Chain Reaction for Molecular Detection of Babesia Felis in Domestic Cats
Muhammad Younus Khan1, Haroon Akbar1, Raheela Akhtar2*, Wasim Shehzad3, Saher Islam3, Beenish Zahid4
1Department of Parasitology, University of Veterinary and Animal Sciences, Lahore, Pakistan.54000; 2Department of Pathology, University of Veterinary and Animal Sciences, Lahore, Pakistan.54000; 3Institute of Biochemistry and Biotechnology (IBBt), University of Veterinary and Animal Sciences, Lahore, Pakistan.54000; 4Department of Zoology, University of the Punjab, Pakistan.
Abstract | The aim of the present study was to optimize the polymerase chain reaction for molecular detection of Babesia felis in domestic cats. One hundred cats suspected for babesiosis with severe signs of pale mucus membrane, haemolytic anaemia, rough body coat and haemoglobinuria were recruited in this study. Polymerase chain reaction was optimised targeting amplification of 18S rRNA gene. Amplification and phylogenetic analysis confirmed Babesia felis in 14% of domestic cats in comparison to microscopy that detected only 6% of cases. PCR also indicated the higher prevalence in local breeds (16.6%) as compares to Persian (13.33%) and Siamese (11.11%). The results indicated that younger (under one year) and older (above three years) cats were more susceptible to infection. There was an increased percentage of Babesia felis in female cats (15%) as compared to male (12.5%). The application of PCR may be recommended for reliable field diagnosis of Babesia felis.
Keywords | Babesia felis, Persian, PCR, Sequencing, Siamese
Editor | Muhammad Imran Rashid, Department of Parasitology, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | April 13, 2018; Accepted | May 06, 2018; Published | June 25, 2018
*Correspondence | Raheela Akhtar, Department of Pathology, University of Veterinary and Animal Sciences, Lahore, Pakistan.54000; Email: raheela.akhtar@uvas.edu.pk
Citation | Khan MY, Akbar H, Akhtar R, Shehzad W, Islam S, Zahid B (2018). Optimization of polymerase chain reaction for molecular detection of babesia felis in domestic cats. J. Adv. Parasitol. 5(2): 22-24.
DOI | http://dx.doi.org/10.17582/journal.jap/2018/5.2.22.24
Copyright © 2018 Khan et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Babesiosis is caused by an intra-erythrocytic haemoprotozoan called Babesia in mammals, birds and reptiles. The economic importance of this disease cannot be neglected due to haemolytic anaemia followed by increased fatality (Laha et al., 2015). Babesiosis is routinely diagnosed through light microscopy when piroplasms are observed in Giemsa-stained thin blood smears. However, it can be difficult to detect Babesia through light microscopy specially if there is chronic infection and when there is low parasitaemia. Drug-resistance associated polymorphism of the parasite may be the cause of false results if the microscopic evaluation is not conducted by experienced personnel (Jiang et al., 2015). In addition antibody detection by enzyme linked immunosorbent assay (ELISA) and immunofluorescent antibody test does not provide direct information about active infection by Babesia (Maharana et al., 2016). In order to prescribe an accurate and suitable treatment for babesiosis, it is necessary to identify both species and genotypes since different infecting species result in various degrees of disease severity and treatment response (Brikenheuer et al., 2003). This highlights the need for molecular diagnosis of Babesia species. As there has not been much work on polymerase chain reaction (PCR) based detection of Babesia felis particularly in Pakistan where not a single study has been conducted for molecular detection of B. felis therefore the present study was designed to optimize PCR for confirmatory diagnosis of B. felis in domestic cats of Lahore, Pakistan.
MATERIAL AND METHODS
One hundred (n = 100) blood samples were collected at Pet Center, University of Veterinary and Animal Sciences, Lahore, Pakistan from domestic cats suspected of babesiosis. The basic criteria for suspecting babesiosis was pale mucus membrane, haemolytic anaemia, rough body coat and haemoglobinuria. Thin blood smears stained with Giemsa were microscopically examined for the detection of intra-erythrocytic bodies. The size of intra-erythrocytic bodies was also determined by using micrometer. Microscopically positive samples were used for optimization of PCR. DNA was isolated using TIANamp Blood DNA kit (Cat # DP318, Tiangen, China).
Primer Design
Optimization of gradient Polymerase Chain Reaction (PCR): The blood samples were also used for detection of Babesia by PCR with specifically designed primers of Babesia against 18S rRNA gene using following primer set:
| Forward | Reverse | |
| B-felis-PB-F | 5’TTATTCGGACTGAGAAACTTAGAT3’ |
5’AGGTTCACAAAACTTCCCAA3’ |
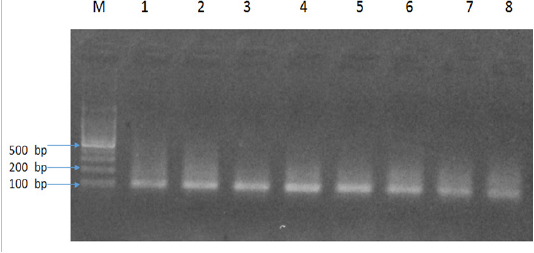
Two (2) µL) of each DNA preparation was added to a 48µL reaction mixture consisting of 1.5U of Taq DNA polymerase (Promega, Madison, WI, USA), 200 mM of each dNTP, 25 pmol of each primer, 5µL 10X PCR buffer, 1.5 mM MgCl2 (Promega, Madison, WI, USA) and remaining water. PCR reaction cycle consisted of an initial denaturation at 94ºC for 10 min, followed by 40 cycles of denaturation at 94ºC for 30s, annealing temperatures in gradient PCR 55-65ºC. Final extension was done at 72ºC for five min followed by a hold step at 4ºC. Using above mentioned primers pair Babesia felis was amplified at size of102bp melting temperature 58ºC.
Sequencing of PCR products and phylogenetic analysis: PCR productswere sequenced using forward primer and aligned by bioinformatic tool (BLAST) and phylogenetic analyses were performed in Mega Align Programme of DNASTAR Laser gene. The data was analyzed using Chi-square (χ2) test.
RESULTS
The results of present study indicated six Babesia felis positive samples by microscopy and 14 by PCR with an amplicon size of 102bp (Figure 1). In addition, there was an increased PCR percentage of Babesia felis in local breeds (16.6%) as compared to Persian (13.33%) and Siamese (11.11%). The present study also determined the age-related prevalence of babesiosis. The cats were divided into three age groups; under 1 year of age, 1-3 years and 4-6 years. PCR results indicated that the highest percentage was found in young cats under 1 year of age (15.38%), followed by 4-6 years (14.28%) and 1-3 years (12.5%). Similarly an increased percentage of Babesia felis was observed in female cats (15%) compared to male (12.5%).
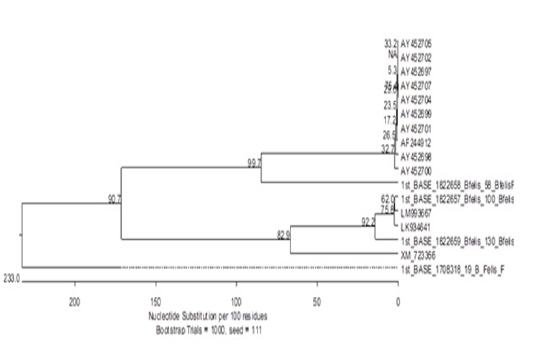
Our results showed some novel findings during phylogenetic analysis when one sequence clustered out with other sequences of Babesia felis from gene bank (Figure 2) while two totally new sequences were found. One of these sequences had about 48% resemblance to Plasmodium yoelii, a species found in mouse. The third sequence had a 69% resemblance with Parascaris equorum. The reasons behind it could be a contamination or remain to be explored.
DISCUSSION
The differential number of positive samples by microscopy and PCR indicates variable detection levels of both techniques. In microscopy, due to its small size the protozoan may easily be skipped. This necessitates the technical skills of microscopy or some alternative technique with more detection ability.
In present study the incidence of positive samples of Babesia felis by microscopy was almost double than the previous studies (Ahmad et al., 2011) conducted at same research site using microscopy. This may be explained by the increasing prevalence of babesiosis. While PCR gave four times more positive samples than reported previously (Ahmad et al. 2011).
The increased incidence in local breeds of cats as compare to is not in line with the previous studies of Ahmad et al. (2011) who found highest prevalence in Siamese and least in Persian. This could again be due to the differential sensitivity of techniques applied (microscopy vs PCR). The increased prevalence of Babesia felis in younger and older cats could be associated with decreased immunity in these age groups.
The presence of Plasmodium yoelii in cats infected with Babesia could be an important finding as it may be a result of a jumping event from one host to another (mouse to cat). The remaining possibilities are mosquitoes that are already well-known intermediate hosts of Plasmodium. This may also re-inforce the hypothesis that cats in Lahore may be harbouring a new species/strain of a haemoparasite. However, it would be pre-mature to say yet. To have more insight into it, we should target more genes with a greater number of amplicons for sequencing and phylogenetic analyses. To the best of our knowledge the present study first time conducted PCR for Babesia felis in Pakistan and due to its more sensitivity and specificity PCR can be recommended as a confirmatory diagnostic tool for detection of Babesia felis.
Acknowledgements
This research was funded by department of Parasitology, UVAS, Pakistan.
Conflict of interest
There is no conflict of interest in this article.
Authors Contribution
MYK and HA designed the study. MYK, RA and SI performed the experiments in lab. WS and BZ performed the statistical analysis. RA drafted the manuscript. All authors read the article before submission.
References