Advances in Animal and Veterinary Sciences
Research Article
Effect of Mycoplasma bovis on Production of Pro-Inflammatory Cytokines by Peripheral Blood Mononuclear Cells
Rekha Valsala1, Rajneesh Rana2*, Arun Thachappully Remesh1, Sabarinath Thankappan3, Sabita Behera1
1Division of Bacteriology & Mycology, IVRI, Izatnagar, Bareilly, UP, India; 2Division of Bacteriology & Mycology, IVRI, Izatnagar, Bareilly, UP, India; 3Division of Bacteriology & Mycology, IVRI, Izatnagar, Bareilly, UP, India.
Abstract | Mycoplasma bovis is an important bovine pathogen associated with calf pneumonia, mastitis, arthritis and sepicemia. It is known to persist in phagocytic and nonphagocytic cells and disseminate to various organs causing systemic infection. In this study, the effect of live M. bovis on pro-inflammatory cytokine production by bovine peripheral blood mononuclear cells was assessed by relative quantification. Temporal change in cytokine levels was assessed at 8 hourly intervals. IL-6 was upregulated (9.32 fold at 24h) by live M. bovis at MOI of 1: 10, while TNF-α levels remained unchanged over time. IL-1 and TNF-α levels were not altered by M. bovis as compared to the control group. This showed that pro-inflammatory cytokines are not much upregulated at low multiplicity of infection.
Keywords | Mycoplasma bovis, IL-1, IL-6, TNF-α, Bovine PBMC
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | June 28, 2017; Accepted | August 22, 2017; Published | September 29, 2017
*Correspondence | Rajneesh Rana, Division of Bacteriology & Mycology, IVRI, Izatnagar, Bareilly, UP, India; Email: rajneeshran01@yahoo.com
Citation | Valsala R, Rana R, Remesh AT, Thankappan S, Behera S (2017). Effect of mycoplasma bovis on production of pro-inflammatory cytokines by peripheral blood mononuclear cells. Adv. Anim. Vet. Sci. 5(10): 400-404.
DOI | http://dx.doi.org/10.17582/journal.aavs/2017/5.10.400.404
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2017 Valsala et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Mycoplasmas are the minimalist microbes, with bare minimum genes as compared to other bacteria and have limited metabolic capability for replication and survival. Even though they are having only half the sets of genes compared to other bacteria, they are pathogenic and are capable of producing variety of diseases in different host species. Besides, many economically important diseases in ruminants like CBPP (Contagious Bovine Pleuropneumonia), CCPP (Contagious Caprine Pleuropneumonia), contagious agalactia, calf pneumonia etc. are caused by mycoplasmas.
Mycoplasma bovis is one of the important agents associated with a number of diseases in bovines, ranging from pneumonia and arthritis in calves to mastitis, genital tract infections, kerato conjunctivitis, otitis media etc. in adult ones (Adamu et al., 2013). Drop in milk production and calf mortality resulting from the disease produces huge economic impact to the dairy industry. The ability of the organism to get adapted to various tissues and to produce diseases indicates the diversity of still unclear pathogenic mechanisms. In many infections, M. bovis acts as an exacerbating agent and often goes undiagnosed or overlooked.
Very little knowledge is available regarding the pathogenic mechanism and virulence factors contributing to mycoplasmal diseases in spite of the small sized genome with limited coding capacity. First step in pathogenesis is adhesion and colonisation (Razin and Jacobs, 1992). Certain adhesins have been identified in mycoplasmas. Mycoplasmal membrane is important in colonization and establishment of infection, as the organism lacks cell wall (Rottem, 2003). Other pathogenic mechanisms include production of hydrogen peroxide and superoxide radicals (Schott et al., 2014). Moreover, membrane lipoproteins, which undergo phase and size variation aid in immune evasion (Rosengarten et al., 2000). Phase and size variation of membrane lipoproteins aid in evading the host immune response (Behrens et al., 1994). A lymphoinhibitory peptide has been identified in M. bovis (Vanden Bush and Rosenbusch, 2004). M. bovis is able to invade and persist in phagocytic as well as non-phagocytic cells, which allows systemic spread of infection without being recognised by host immune system (Burki et al., 2015).
Immune response contributes to the pulmonary tissue damage characteristic of mycoplasmal respiratory infections (Van den Bush, 2003). Interaction of mycoplasma with host immune cells results in production of pro-inflammatory cytokines as well as chemokines. The production of nitric oxide and TNF-α are induced in alveolar macrophages on co-incubation with M. bovis (Jungi et al., 1996). Besides bovine γδ T-cells are activated, which are also part of innate immunity. Van der Merwe et al. (2010) showed persistence of M. bovis in bovine peripheral blood mononuclear cells and erythrocytes. This study was conducted to evaluate the effect of M. bovis on production of pro-inflammatory cytokines by bovine PBMCs.
RESULTS AND DISCUSSION
PBMCs were isolated from cattle blood by density gradient centrifugation. The cell count in bovine PBMC was 1-1.225 x 106 cells/ml and viability percentage was 91.49-93.02%. RNA was extracted from PBMC samples after co-incubation for different time periods with live M. bovis. On analysis of purity and concentration of RNA samples by nanodrop, all the samples were having A260/A280 between 1.8-2.0, indicating purity and concentration ranged from 500-1000 ng/µl. cDNA was synthesized from all the RNA samples using Revert Aid First Strand cDNA synthesis kit.
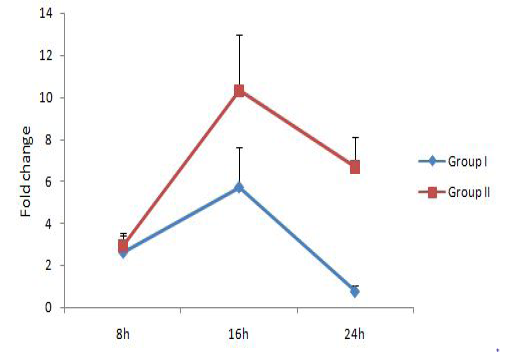
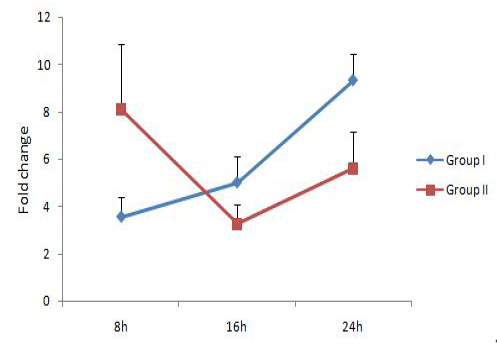
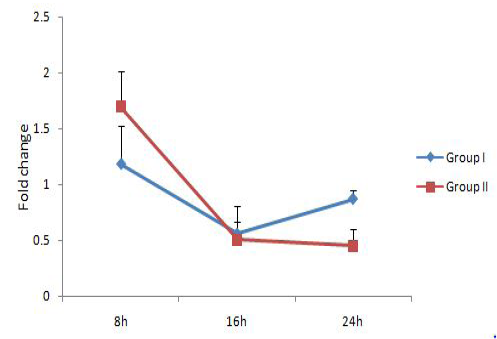
The relative expression of IL-1, IL-6 and TNF-α mRNA were calculated in terms of fold change with respect to housekeeping gene YWHAZ. YWHAZ codes for Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide and was earlier reported to be a suitable reference gene for bovine samples (Spalenza et al., 2011). Previously reported primers which bind at the exon-exon junctions were used in this study (Yamada et al., 2009; Puech et al., 2015). Group I represents bovine PBMC treated with live M. bovis at MOI of 100:1 and group II represents untreated control. Results are represented in Figure 1, 2 and 3 respectively for IL-1, IL-6 and TNF-α.IL-1 expression level was similar in both groups at 8h, followed by a significant upregulation at 16 h. At 24h, the IL-1 expression level reduced in both the groups, but the level was lower in group treated with M. bovis. This result indicated that M. bovis co-infection resulted in a comparatively lower IL-1 response. Reduced IL-1 mRNA
expression level has been demonstrated previously in squamous cell carcinoma A431 cells infected with mycoplasmas as compared to non-infected cells (Demczuk et al., 1988). IL-6 gene expression level was showing an increasing trend over time and was significantly upregulated in group I (9.32 fold) at 24h. But the expression level reduced in group II at 16h as compared to 8h followed by an increase at 24h.TNF-α level was not showing any significant changes with time. These results showed that M. bovisco-infection resulted in upregulation of the pro-inflammatory cytokine IL-6 alone and a comparatively lower IL-1 response.
Inflammatory cytokines are part of body’s immediate defense response to ‘danger’ and immune response is greatly enhanced by them (Murtaugh and Foss, 2002). They may play an important role in enhancing the biological response of bovine neutrophils to bacterial infections (Leite et al., 2002; Sohn et al., 2007). Rodriguez et al. (2015) conducted immunohisto chemical study for cytokine expression in spontaneously infected animals and found out that M. bovis-associated pneumonic lesions consistently resulted in upregulation of TNF-α, IL-4, IL-10 and IFN-γ expression, but expression of IL-1α, IL-1β, IL-2, IL-6 and IL-8 did not exhibit significant differences between infected and normal lung from control calves. In a similar study by Gondaira et al. (2015) live M. bovis was shown to significantly induce the production of tumor necrosis factor alpha (TNF-α), interleukin 12p40 (IL-12), and interferon gamma (IFN-γ) mRNA expression in bovine PBMCs at a multiplicity of infection (MOI) of 1000 but not at MOI of 10 and 100. The relatively lower inflammatory cytokine response in our study might be due to the low MOI.
Alveolar macrophages are activated by M. bovis and produce TNF-α and nitric oxide (Jungi et al., 1996). This is important in pathogenesis as well. Jimbo et al. (2017) showed in vitro production of TNF-α and IL-12 by neutrophils on infection with M. bovis priming to Th1 response. In monocytes, M. bovis delays apoptosis, and suppresses production of IFN-γ and TNF-α (Mulongo et al., 2014), which aids in persistence and systemic dissemination of M. bovis by effective immunomodulation.
Production of pro-inflammatory cytokines is important with respect to pathogenesis of M. bovis. Although there was an upregulation of IL-6 on co-incubation of PBMCs with live M. bovis at an MOI of 100:1, the levels of IL-1 and TNF- α were not increased. It can be concluded that it is required to have high multiplicity of infection for producing a consistent inflammatory response.
MATERIALS AND METHODS
Mycoplasma bovis Standard strain NCTC 10131 was obtained from Referral Laboratory on Mycoplasma, Division of Bacteriology and Mycology, Indian Veterinary Research Institute, Izatnagar (U.P) and was grown in modified PPLO broth medium (PPLO broth, yeast extract, glucose, sodium pyruvate, phenol red 0.5%, thallous acetate 5%, penicillin, DNA 0.2%, horse serum 10%). Pelleted the cells by refrigerated centrifugation at 10000xg for 15 min. After one was step with phosphate buffered saline (PBS, pH 7.4) M. bovis was resuspended in PBS. The concentration was adjusted to approximately 108 cells/ml.
Isolation of Bovine Peripheral Blood Mononuclear Cells (Pbmcs)
Blood (10 ml) was collected in anticoagulant ACD-A containing tubes from 3 healthy animals and was used for PBMC isolation by gradient centrifugation over histopaque 1083 (Sigma Aldrich) following the manufacturer’s protocol. PBMCs were resuspended in RPMI-1640 medium containing horse serum and antibiotics. Viable cell count was determined by counting in haemocytometer counting chamber following trypan blue staining.
Co-Incubation with Live M. Bovis
PBMC concentration was adjusted to 2 x 106 cells/ ml and 100 μl each was added to wells in a 96 well tissue culture plate. In one row, M. bovis was added at an MOI of100:1. Cells from each animal were plated in triplicate. The cells were incubated in 3 different plates at 370C, 5% CO2. One plate each was removed at 8h intervals.
Total RNA Extraction
Total RNA extraction from the PBMC samples collected was done by Ribozol method. The concentration of RNA was checked by Nanodrop (NanoVue Plus, GE Lifesciences). RNA purity was determined by 260:280 ratio. Total RNA with the ratio values 1.8 or greater are deemed acceptable for subsequent procedure and was stored at -70ºC.
Reverse Transcription for Cdna Synthesis
Synthesis of cDNA from total RNA was carried out by using Revert Aid First Strand cDNA synthesis kit (Thermoscientific, U.S.A.). In a nuclease free tube 1000 ng of total RNA was taken and to this 1µl of oligo(dT) primer (100µM) was added. Total volume was adjusted to 12 µl using nuclease free water. The contents were gently mixed and incubated at 65oC for 5 minutes. The tube was then chilled on ice followed by addition of 4µl 5X reaction buffer, 1 µl of RiboLock RNase Inhibitor (20 U/µl), 2 µl of 10 mM dNTP Mix, and 1 µl of RevertAid M-MuLV Reverse Transcriptase (200 U/µl) to get a final volume of 20 µl. The reaction mixture was mixed and centrifuged briefly and then incubated at 42oC for 60 minutes. The reaction was terminated by heating at 70oC for 5 minutes. Finally the product was stored at -20oC.
Pro-Inflammatory Cytokine Expression Study by Real Time Pcr
Study of differential expression of 3 cytokine genes IL-1, 1L-6 and TNF-αm RNA from PBMC samples was done by relative quantification. Real-time PCR was carried out using YWHAZ as reference housekeeping gene (endogenous control). Primers used for real time PCR are given in Table 1. All the primers were procured from Eurofins MWG Operon, Bangalore. Reaction was carried out in a total volume of 15μl. For each sample, a non-template control (NTC) for both target and endogenous genes, which included all ingredients of real time reaction except the template, was run to check contamination of any ingredient.
Table 1: Real time PCR primers
| Gene | Primer (5’-3’) |
| IL-1 | CCTTGGGTATCAGGGACAA |
| GGGTATGGCTTTCTTTAGG | |
| IL-6 | TGAGTGTGAAAGCAGCAAGGA |
| TACTCCAGAAGACCAGCAGTGG | |
| TNF-α | CCAGAGGGAAGAGCAGTCC |
| GGCTACAACGTGGGCTACC | |
| YWHAZ | GAAAGGGATTGTGGACCAG |
| GGCTTCATCAAATGCTGTCT |
Real-time PCR was performed on a CFX96 C1000 Touch QPCR System (Biorad, U.S.A.) using Kapa SYBR Green mastermix (Sigma Aldrich, U.S.A.). Samples were run in duplicates and relative expression was studied. Three stage PCR protocol was set for the real-time amplification of cDNA for each and every sample. First stage consisted of initial melting of template for 3 minutes at 95°C with no repetition. Second stage consisted of 2 steps repeated forty times with first step of melting template at 95°C for 10 seconds and second step of annealing/extension at 60oC for 30 seconds except for IL-6. For IL-6, annealing was at 540C for 15sec followed by an extension step at 720C for15 sec. Third stage consisted of three steps and is carried out for drawing the dissociation curve of products generated during second stage.
After 40 cycles of amplification, threshold cycle (Ct) value was obtained for PBMC cytokines and internal control gene YWHAZ (reference gene). Melting curve analysis was performed for each sample to verify the specificity of each product. The fluorescent data acquisition was continuous from annealing to the final denaturation step. The Real time data obtained was analyzed by CFX96 Touch QPCR Software (Biorad, U.S.A.). Ct values were used to determine the relative quantification of cytokines.
The change in the level of expression of PBMC cytokine mRNA was calculated by using the ΔΔCt method. The statistical significance of differences in mRNA expressions of the examined factors was assessed by one way ANOVA test using SPSS 20.0 software. Differences were considered significant if P<0.05.
ACKNOWLEDGEMENTs
Authors are thankful to the Director, Indian Veterinary Research Institute, Izatnagar for providing the necessary facilities to carry out the experiment. First author gratefully acknowledges the receipt of junior research fellowship by University Grants Commission, New Delhi, India during the research tenure.
Conflict of interest
The authors declare that they have no conflicts of interests.
Authors Contribution
All the authors have contributed in terms of technical knowledge in framing the article.
REFERENCES