Advances in Animal and Veterinary Sciences
Research Article
Molecular Identification of Candida Species Isolated from Ears of Dogs Infected with Otitis externa by Detecting Internal Transcript Spacer (ITS1 and ITS4) in Sulaimania, Iraq
Hanan Hasan Ali1*, Rana Mohamed Al-Obaidi2, Chiman Hamarahim Fattah1
1Department of Microbiology; 2Department of Basic Science, School of Veterinary Medicine, University of Sulaimania, Sulaimania, Iraq.
Abstract | The goal of this study was to determine internal transcribed spacer/5.8S ribosomal DNA (rDNA) to detect Candida spp. in dogs with Otitis externa. PCR-based detection of internal transcribed spacer 2 (ITS2) regions of the rRNA genes was evaluated as a means of fungal identification by using Internal transcribed spacers 1 (ITS1) and Internal transcribed spacers 4 (ITS4). These were amplified by PCR to detect fungal DNA. Sixty swab ear samples from domestic dogs with and without O. externa were examined for clinical signs including head shaking, scratching the external ear flaps, bad odor, obstraction of the ear canal, anorexia, emaciation and auricular discharge (unilateral in 20 and bilateral in 10 dogs). The isolation of Candida species was performed from the external otitic canal in all animals. Yeast species isolated in 30 cases involving mainly Candida spp. (20 cases) and more often Aspergillus niger (5 cases), Penicellium spp. (3 cases) and Aspergillus fumigatus (2 cases). Twenty isolates of Candida used to test the selected primers and conditions of the PCR. Molecular fungal identification is successful in most isolates; with a band size 600-800 bp. This study concluded that size of PCR-amplified ITS2 region DNA is a rapid and reliable method to identify clinically significant yeasts.
Keywords | Candida spp, Candida famata, Candida kefyr, Otitis externa, Dogs, Colony PCR
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | June 27, 2015; Revised | July 17, 2015; Accepted | July 19, 2015; Published | July 29, 2015
*Correspondence | Hanan Hasan Ali, University of Sulaimania, Sulaimania, Iraq; Email: dr.hananh23@gmail.com
Citation | Ali HH, Al-Obaidi RM, Fattah CH (2015). Molecular identification of Candida species isolated from ears of dogs infected with Otitis externa by detecting internal transcript spacer (ITS1 and ITS4) in Sulaimania, Iraq. Adv. Anim. Vet. Sci. 3(9): 491-499.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.9.491.499
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Ali et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Candida is a genus of yeasts and is the most common cause of fungal infections worldwide. The virulence of Candida isolates differs according to the species; many species are harmless commensals or endosymbionts of hosts including humans. Detection and identification have been made even more important because of the emergence of candidemia caused by innately fluconazole-resistant non-Candida albicans Candida species (Fujita et al., 2001). However, when mucosal barriers are disrupted or the immune system is compromised they can invade and cause disease. C. albicans is the most commonly isolated species, and can cause infections (candidiasis or thrush) in humans and other animals. In winemaking, some species of Candida can potentially spoil wines (Manolakaki et al., 2010).
Candida spp. is a natural inhabitant of the mucous membranes of the genital, alimentary and upper respiratory tract of animals (Greene and Chandler, 1998). This opportunistic yeast-like pathogen can cause localized infection in immunosuppressed patients, most often in those receiving long-term corticosteroid therapy, prolonged antimicrobial therapy, cytotoxic chemotherapy and patients with diabetes mellitus (Jones et al., 1997). Systemic candidiasis is rare in animals and there are scant reports in the literature describing multisystemic infection (Rodriguez et al., 1998; Heseltine et al., 2003; Brown et al., 2005; Kuwamura et al., 2006).
Diseases of the external ear are commonly encountered in dogs and are among the most frustrating in diagnostic and therapeutic point of view to practicing veterinarians. In the absence of detail observations, diagnostic tools, most of the canine practitioners, frequently initiate treatment of O. externa prior or even without sensitivity testing.
O. externa can cause considerable pain, distress and discomfort to the affected dog. The difficulty in treating O. externa is due to the complexity and multiple etiological agents and emergence of drug resistance. The etiological components for auricular disorders are vast and involve several factors and its identification may be the key for formulating successful therapy. Relapse or recurrence of ear infection is often noticed in O. externa dogs even with symptomatic treatment (Lakshmi and Tirumala, 2013).
O. externa is an acute or chronic inflammation of the external ear. The disease is more frequent in dogs than in cats. Dogs with long pendulous ears are most commonly affected. Ear infections are among the ten most frequent reasons for dogs to be presented to veterinarians and may affect up to 20% of dogs (Angus, 2004), Candida spp. also progressing pathological alterations as hyperplasia, oedema, fibrosis (Greene, 2006).
The virulence of Candida isolates differs according to the species, with C. albicans being most virulent, followed by Candida tropicalis (Wingard, 1995); thus, the prompt and accurate detection and identification of yeast species are very important.
Various commercial systems that can identify these pathogens within 4 to 72 h have been developed (Espinel-Ingroff et al., 1998). Although correct identification of clinically relevant yeast strains can be achieved with these systems, incomplete or incorrect identification may occur when certain new and emerging yeast strains are tested (Pfaller et al., 1996). Moreover, culture-based phenotypic identification of Candida species from clinical materials requires at least 1 day obtaining a pure culture.
In recent years, numerous DNA-based methods have been developed to improve the diagnosis of mycotic infections and the identification of pathogenic fungi. Polymerase chain reaction methods are particularly promising because of their simplicity, specificity, and sensitivity (Lou and Mitchell, 2002).
PCR-based techniques have contributed to the identification of fungal species from clinical specimens. The PCR technique is commonly used for identifying Candida species (Alothman, 2012). In recent years novel molecular methods, notably the amplification of gene sequences unique to fungi by PCR assays, have been developed to improve the diagnosis of life-threating invasive fungal infections in high risk patients. PCR offers the potential for rapid diagnosis (Peman and Zaragoza, 2010), thereby enabling the possibility of earlier diagnosis and more timely initiation of antifungal therapy, when appropriate (Ostrosky-Zeichner, 2012). The ITS region is the most widely sequenced DNA region in molecular ecology of fungi (Peay et al., 2008) and has been recommended as the universal fungal barcode sequence (Schoch et al., 2012). It has typically been most useful for molecular systematics at the species level, and even within species (e.g., to identify geographic races). Because of its higher degree of variation than other genetic regions of rDNA (for small- and large-subunit rRNA), variation among individual rDNA repeats can sometimes be observed within both the ITS and IGS regions. Turenne et al. (1999) reported that differences in the sizes of the ITS2 regions of fungi were useful for the rapid identification of clinically important fungi. In addition to the standard ITS1+ITS4 primers used by most labs, several taxon-specific primers have been described that allow selective amplification of fungal sequences (White et al., 1990).
This study aimed to isolate pathogenic Candida spp. from dogs with O. externa infection and identification of the pathogen by routine Mycological, biochemical tests and molecular typing by using PCR as a rapid confirmatory test For identification the main Candida spp.
MATERIALS AND METHODS
Clinical Examination and Specimen Collection
Complete physical and dermatological examinations were performed prior to collection of O. externa specimens in each dog. The following data were recorded for each subject: age, sex, breed and ear type (erect or pendulous). Physical examination findings were categorized as normal or consistent with allergic skin disease. Findings consistent with allergic skin disease included one or more of the following: inter-digital erythema, salivary staining, barbering, hypotrichosis and exco-riations. Additionally, pustules, epidermal collarettes, lichenification and post-inflammatory hyperpigmentation were considered signs of secondary pyoderma due to underlying hypersensitivity and thus were included as consistent with allergic skin disease.
Animals and Sampling Collection
The study included 60 dogs from both genders, from three weeks to three years old. According to the clinical examinations the animals were divided equally into two groups, the first group was included dogs with signs of the O. externa, whereas dogs of the second group were included clinically normal dogs. Infected animals were presented with unilateral or bilateral dropping of ears, head shaking, pruritus, pain when palpated, erythema and swelling of ear skin or of the ear canal with increased amount of cerumen. Samples were collected using sterile cotton swabs which directly enters the external ear. The study was performed between December 2013 and January 2014 in Sulimania province.
Table 1 show that the total fungal infection proportion of group with O. externa (70%) and without O. externa (30%) differed significantly (P < 0.01). These results is consistence with results obtained by Sapierzyński (2009) confirmed the presence of the fungi in 68% of the affected dogs, whereas Mansfield et al. (1999), Ziółkowska and Nowakiewicz (2004) as well as Bond et al. (1995) found in their studies on O. externa that in 80–83% of the examined dogs the disease was caused by Malassezia fungi.
Table 1: The percentages of infected swabs from external ear canal in dogs
|
Animal Groups |
Numbers of examined swabs |
Numbers of positive isolates |
|
With Otitis externa |
30 |
21(70%) |
|
Without Otitis externa |
30 |
9(30%) |
|
Chi square |
9.60 |
|
|
P |
< 0.01 |
Results showed that the difference in fungal infection proportion between with and without O. externa for all type of fungi was not significant except Gandida spp. On the other hand, the differences among all type of fungi with O. externa were significant (P < 0.01). The highest proportion of infection was detected in Gandida spp. (50%), whereas the differences among all type of fungi were not significant in without O. externa.
Isolation of Fungal Cultures Swabs
Different types of fungal was isolated from O. externa dogs which have main clinical signs or without (Table 2 and Figure 2, 3, 4), yeast and mould by culturing on sterilizing Sabouroud Dextrose Agar and adding antibiotic chloramphenicol and Cycloheximid for inhibit the growth of non-pathogenic fungi and incubated at different temperature 30-37ºC for yeast and 25-30ºC for mould for (3-5) days, Then prepare direct slide culture and staining by Lacto phenol cotton blue.
Isolation and Identification of Candida spp.
Macroscopic: Candida grows rapidly in culture, reaching maturity in as little as three days at 30- 37ºC. Colonies were cream colored, raised, entire, smooth and butyrous on Sabouroud dextrose agar, the colonies may develop small striations or outgrowths often referred to as “feet” which are indicative of some Candida species (Figure 2). Identification of Candida was performed according to the method of (Haley, 1971) as followed: i. Germ tube which is considered as the specific test for identification of the Candida; ii. Microscopically; and iii. Grossely.
CHROMagar Candida medium: It is a selective and differential medium for the isolation of fungi. With the inclusion of chromogenic substrates in the medium, the colonies of different Candida species produce different colours, thus allowing the direct detection of these yeast species on the isolation plate. The specimens were streaked for isolation onto the surface of the medium. Plates Incubated aerobically at (35-37)ºC for 20 to 48 hours in an inverted position. An incubation time of 42 hours was required for the full colour development of Candida colonies.
Colony PCR: Single Candida colony was directly used as template for PCR without isolating pure DNA, by colony polymerase chain reaction (colony PCR). Through colony PCR, the large amount of material and time-consuming process involving extraction of genomic DNA can be saved. A single colony was picked up from overnight culture and re-suspended in 50µl dH2O. The DNA was released by incubation at 95ºC for 15min (Akan, 2006; Zhong, 2011).
Detection of Candida spp. by PCR: The PCR designed by Chang et al. (2001) was used to amplify intergenic spacer regions (ITS) of ribosomal DNA (rDNA) with the primers
Table 2: Different fungal isolated from dogs with and without Otitis externa
|
With Otitis externa |
Without Otitis externa |
||||||||
|
Fungal Infection |
No. |
Single Isolation |
Mixed Isolation |
% |
Single Isolation |
Mixed Isolation |
% |
Chi sq. |
P |
|
Candida spp. |
30 |
12 |
3 |
50 |
3 |
2 |
16.66 |
7.50 |
<0.01 |
|
A. fumigatus |
30 |
0 |
2 |
6.66 |
0 |
0 |
0 |
2.06 |
0.15 |
|
A. niger |
30 |
2 |
0 |
6.66 |
0 |
3 |
10 |
0.21 |
0.64 |
|
Penicellim Spp. |
30 |
0 |
2 |
6.66 |
0 |
1 |
3.33 |
0.35 |
0.55 |
|
Chi sq. |
29.26 |
7.08 |
|||||||
|
P |
<0.01 |
0.07 |
|||||||
ITS1(5’-TCCGTAGGTGAACCTGCG-3’), and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’), targeting the conserved regions of 18S, 5.8S, and 28S rDNA (White et al., 1990), respectively, were used for amplification. The ITS1-ITS4 primer pair was used to amplify the intervening 5.8S rDNA and the adjacent ITS2 and ITS3 regions (Figure 5).
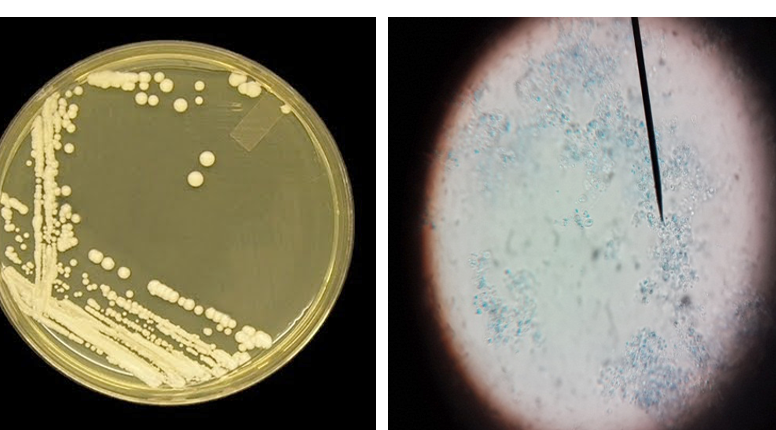
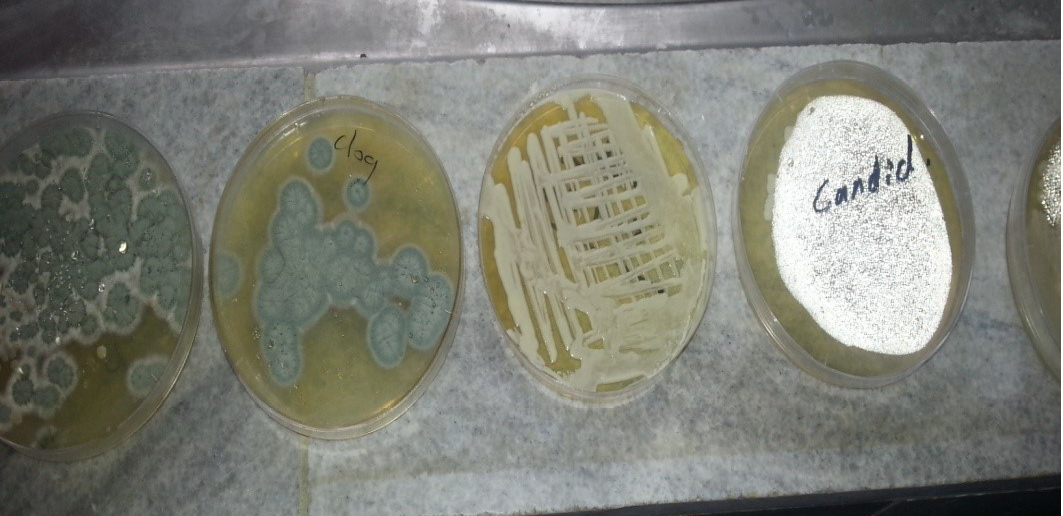
Figure 2: Candida colonies creamy muciod appear on SDA and Malt extract on left side and, unicellular cell of Candida stained by Lacto phenol cotton blue under X 400 on right side
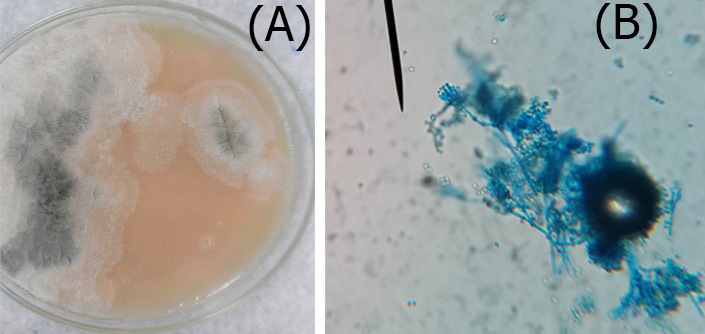
Figure 3: A. Colonies of Penicillium on SDA; B. Penicillium spp. showing brush like arrangement of fruiting head of the conidiophores microscopically
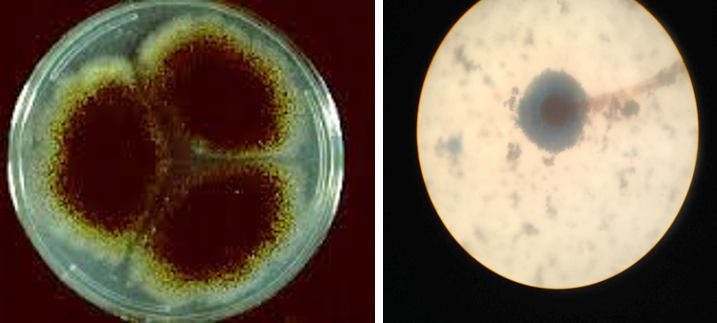
Figure 4: Colony of Aspergillus niger (Black colony) on SDA, PDA from Site of infection and Aspergillus niger isolated from A
For the optimum PCR conditions, a reaction volume of 100µl contained 0.2 mM each deoxyneocleoside triphosphate, 1.5mM magnesium chloride, 0.5µM each primer, 10x Taq buffer and 2.5U of Taq polymerase (DNAmp, England) and 0.5µg of candidal DNA as template were used. Negative controls were performed with sterile deionized water in place of the template DNA. Reaction mixtures were subjected to 35 cycles of the following incubations: Denaturation at 94ºC for 30 sec, primer annealing at 59ºC for 1 min and extension at 72ºC for 45 sec (Chang et al., 2001). A hybrid thermal cycler was used (Bioneer). Ten µl from the amplicons were analysed in 1.5% agarose gel with 1x TBE buffer stained with ethidium bromide and visualized by illumination with UV light (Figure 6).
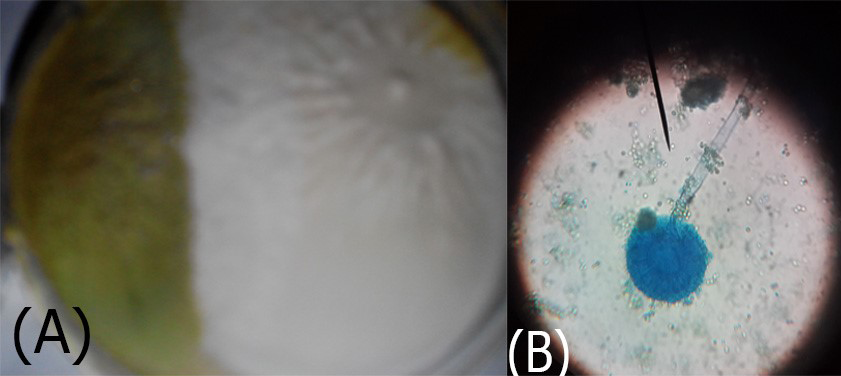
Figure 5: A. Aspergillus flavus colonies greenish-oily colour; B. reproductive head on Conidiophores very clear
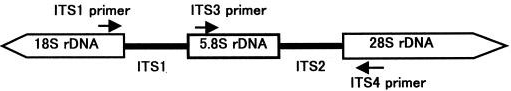
Figure 6: Schematic representation of the fungal ribosomal genes containing the primer target areas used in this study
For the identification of yeasts, various methods are available, including chromogenic substrates (Odds and Bernaerts, 1994), micromorphology on rice agar, and biochemical characterizations. For the latter, several products are commercially available, such as Auxacolor (Bio-Rad), Vitek and API ID32C (bioMérieux).
Nada et al. (2014) have mentioned that the genus Candida includes over 50 species of yeasts, many of which are associated with infections in humans and animals. C. albicans, the only phospholipase-producing species, is worldwide the most frequently isolated from both humans and animals. However, the past decades have demonstrated an increase in the number of none-Albicans species – Candida krusei, Candida parapsilosis, C. famata, Candida kefyr, etc. Moreover, such none-Albicans species are less susceptible to azole derivatives.
Yeast species were isolated in 30 of cases 20 isolates involving mainly Candida spp. (13 isolates of C. kefyr and 7 isolates of C. famata) shown in Figure 1 and more often Aspergillus niger (5 isolates), Penicellim Spp. (3 isolates), and Aspergillus Fumigatus (2 isolates) as shown below in Table 1 and Table 2.
Germ tube test gave negative result to both C. famata and C. kefyr which agree with the results obtained by (Buzzini and Martini, 2001).
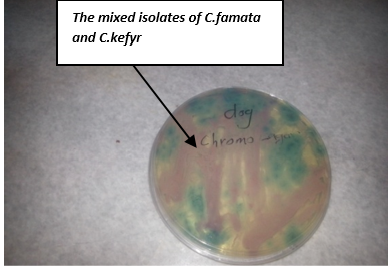
On chromoagagr medium colonies of C. famata appeared white to light pink, while C. kefyr colonies appeared pink to purple (Figure 7). Other yeast species may develop either their natural colour (cream) or appear rose or light to dark mauve (Odds and Bernaerts, 1994). An additional advantage of the medium is the easy detection of mixed yeast cultures due to their colony appearance in different colours (Odds and Bernaerts, 1994; Bauters and Nelis, 2002; Murray et al., 2003).
Willinger and Manafi (1999) suggested that CHROMagar Candida is a differential culture medium that allows selective isolation of yeasts and simultaneously identifies colonies of C. albicans, Candida glabrata, C. tropicalis and C. krusei but it’s not specific for the identification of other Candida species which may be confused because most species of Candida may not grow or give the creamy white or light pink.
In a recent study (Nada et al., 2014) isolated Candida spp. Between July 2010 and February 2012, where 58 Candida Spp. strains were isolated from different sources, such as: milk from cows with mastitis, ear secretions and pharyngeal exudates from dogs, and faeces from parrots, pigeons and chickens.
C. kefyr is the new name of Kluyveromyces marxianus which is a species of yeast in the genus Kluyveromyces, and is the sexual form (teleomorph) of C. kefyr. and K. marxianus, used commercially to produce the lactase enzyme similar to the use of other fungi such as those in the genus Aspergillus. It is produced as a nutritional yeast and bonding agent for fodder and pet food, and as a source of ribonucleic acid in pharmaceuticals, which may explain its presence in many species of animals including dogs and horses (Seyis and Aksoz, 2004).
Other yeasts, such as C. famata, are being recognized as potential emerging pathogens that cause several types of infections in humans and animals. Consequently, we have investigated the adhesion and internalization of C. famata into monocytes and epithelial cells (Pacheco et al., 2007).
In recent years, several DNA-based molecular identification methods have been established which make use of the variable domains of the 18S or 28S rRNA gene (Makimura et al., 1994; Kurtzman and Robnett, 1997; Alothman, 2012).
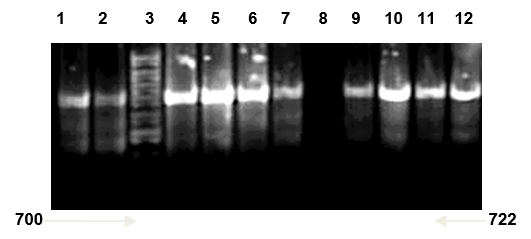
Multiplex PCR was used to further confirmation of isolated Candida spp. types Universal primers ITS1 and ITS4 amplified ITS region was used for investigating 20 clinical isolated Candida spp. successfully. And it yielded a unique PCR product size of approximately 633-722 bp which represent the PCR sizes of C. famata and C. kefyr respectively as shown in Figure 1, 2 and 3, these results agreed with results of (Fujita et al., 2001; Cirak et al., 2003; Pinto et al., 2004; Ciardo et al., 2006; Shokohi et al., 2010) while, the amplification profiles of the PCR products targeting the ITS1-ITS4 region in a recent study by Nada et al. (2014) were C. albicans (535 bp), C. parapsilosis (520 bp), C. krusei (510 bp), C. famata (490 bp), and C. kefyr (840 bp) which disagree with results of the current study (Results shown in Figure 8 and 9).
Candida kefyr lane 1, 2, 4, 5, 6, 7, 9, 10, 11 and 12: band size (722bp); Lane 3: ladder 1kb; Lane 8: negative control (TBE and loading buffer)
Lane 1: DNA ladder 1kb; Lane 2, 3, 4, 5: Candida famata band size (633bp); Lane 6 negative control (TBE and loading buffer)
Furthermore, most isolated species were identified successfully. C. kefyr was the most common species 65% (13/20) while C. famata had the least percentage of 35 (7/20) of the total sample investigated. These proportions disagreed with previous studies (Fujita et al., 2001; Ciardo et al., 2006) The results also differed with the results of Pfaller et al. (2007) who reported that The most of Candida spp. that are involved in invasive candidiasis included C. albicans (62%), C. glabrata (12%), C. tropicalis (7.5%), C. parapsilosis (7.3%), C. krusei (2.7%), C. guilliermondii (0.8%) and C. lusitaniae (0.6%). These studies included samples of human, and because there isn’t any study previously mentioned detecting Candida isolated from ears of dogs by PCR the current study could be the first.
The ITS region, located between the 18S and 28S rRNA genes, is more promising for species discrimination because of its higher variability (Iwen et al., 2001). Although attempts to identify fungi by focusing on either the ITS1 or the ITS2 region may be successful for some species and genera (Chen et al., 2000; Chen et al., 2001), analysis of the complete ITS region offers greater promise for molecular identification.
Various molecular approaches have been used for the detection of fungi from clinical samples, targets for the detection of fungi at the genus or species level have included 18S rDNA (Makimura et al., 1994), mitochondrial DNA (Gardes et al., 1991), intergenic spacer regions (Radford et al., 1998), and ITS regions (Jackson et al., 1999). The sizes of the 18S, 5.8S, and 28S rRNA genes are essentially identical in all species, while the lengths of the ITS regions depend on the species (Peman and Zaragoza, 2010).
As for rapid sizing of PCR products, Turenne et al. (1999), used a capillary electrophoresis system which needs less than 30 min for analysis. Recently, electrophoretic analysis with a microchip was developed (Cheng et al., 1998).
Nguyen et al., (1996) have used PCR- AGE and PCR-ME and reported that the major limitation of PCR-based method for the direct detection of fungi from clinical samples is mixed flora. Mixed candidiasis, have frequency of 3.3% (14 of 427 patients) because PCR-based method with universal primers amplified all kinds of genomic DNA from fungal strains; the use of PCR may be limited. They also mentioned in a preliminary study, electrophoresis of PCR products obtained from a mixture of C. albicans and C. glabrata cultures showed four fragments, indicating mixed flora. Therefore, when four or more fragments are detected with the PCR-AGE or PCR-ME method, the presence of mixed flora should be taken into consideration and the identification of fungi may be difficult.
Nowadays, the opportunistic pathogen Candida causes a life-threatening infection especially in immuno-compromised patients (Neppelenbroek et al., 2005). In spite of sanitary cares and treatment methods, incidence of candidiasis has increased markedly (Mirhendi et al., 2006).
The reason for such dramatic variation in the frequency of Candida spp. in patients are unclear but may include exposure to azoles, age, underlying disease, geographic location, or other, unknown, factors (Pfaller et al., 2007). Since, the virulence of the Candida spp. isolated from different locations is varied, rapid and reliable identification methods are crucial for efficient antifungal treatments.
PCR methods can detect extremely small quantities of DNA and cause earlier detection of pathogenic fungi and consequently allow earlier beginning of antifungal therapy that may improve chances of survival. These methods can directly detect the presence of fungi with high degree of specificity and sensitivity (Mirhendi and Makimura, 2003).
The ITS region is now perhaps the most widely sequenced DNA region in fungi. It has typically been most useful for molecular systematics at the species level, and even within species (e.g., to identify geographic races). Because of its higher degree of variation than other genic regions of rDNA (SSU and LSU), variation among individual rDNA repeats can sometimes be observed within both the ITS and IGS regions. In addition to the standard ITS1+ITS4 primers used by most labs, everal taxon-specific primers have been described that allow selective amplification of fungal sequences (Ciardo et al., 2006).
Pinto et al. (2004) used universal primers ITS1 and ITS4 for the amplification of ITS1 and ITS2 regions, including 5.8S subunit genes and digested it with eight restriction enzymes. They could easily identify Candida spp. on the basis of size and number of bands using the universal primers ITS1 and ITS4 to amplify the ITS1 and ITS4 region and 5.8S in the rDNA gene. These primers have already demonstrated their efficiency (Mirhendi and Makimura, 2005; Pinto et al., 2004). In fact, they could amplify the complete part of ITS1, ITS4 and 5.8S rDNA regions and of 18S and 28S rDNA and too many small fragments about 20-30 bp.
Similar to other papers (Mirhendi and Makimura, 2005; Pinto et al., 2004) the same results were obtained by CHROMagar and PCR methods in detecting different Candida spp., however in our study a couple of isolate showed different results using CHROMagar due to undetermined colour. This finding is a conclusive reason to apply molecular methods for determination and identification of medically important Candida spp. in clinical laboratory.
Since there weren’t any other studies related to the identification of ITS region in Candida species isolated from dogs, thus this study was the first to identify the isolates of Candida isolated from clinical cases of dogs infected with O. externa by PCR in Sulaimania province.
In summary, this method for the identification of clinically relevant fungal isolates is sensitive, rapid, and specific for all yeast organisms tested. PCR analysis is a rapid test in processing yeast-positive blood culture bottles as well as identifying problematic isolates of yeasts in routine laboratory work.
ACKNOWLEDGEMENTS
The authors declare that there is no conflict of interest.
CONFLICT OF INTERESTS
The authors are grateful to the Department of Microbiology, School of Veterinary Medicine and College of Medicine, University of Sulaimania, Iraq for providing the facilities to carry out this work.
AUTHORS CONTRIBUTION
Hanan Hasan Ali designed the study, isolated Candida of the present study, facilitated the study by arranging samples and finalised the manuscript. Rana Mohamed Al-Obaidi, and Chiman Hamarahim Fattah carried out the laboratory work.
REFERENCES