Advances in Animal and Veterinary Sciences
Research Article
Isolation and Characterization of Listeria spp. from Organized and Migratory Sheep Flocks in India
Shivaramu Keelara1*, Satya Veer Singh Malik2, Shivasharanappa Nayakvadi3, Samir Das2, Sukhadeo Baliram Barbuddhe4
1Department of Nutrition and Food Science, University of Maryland, College Park, MD 20742, USA; 2Division of Veterinary Public Health, Indian Veterinary Research Institute, Izatnagar, 243122, India; 3Animal Sciences Section, Central Coastal Agricultural Research Institute, Goa, 403402, India; 4National Institute of Biotic Stresses Management, Raipur, Chhattisgarh, India.
Abstract | In the present investigation, a total of 880 clinical samples comprising blood (n=215), vaginal swabs (n=220), fecal swabs (n=220), placenta (n=10) and sera (n=215) from ewes with abortion or history of abortion as well as from apparently healthy ewes of organized and migratory flocks were collected for screening listeric infection by isolation, serological and molecular diagnostic methods. 23 different isolates were recovered which comprised of 15 L. monocytogenes, 2 L. ivanovii and 6 other listeriae. Among these, four haemolytic isolates (L. ivanovii-2 and L. monocytogenes-2) were found pathogenic based on hemolysis on sheep blood agar, CAMP test, PI-PLC activity, virulence-associated genes (prfA, plcA, actA, hlyA and iap) as well as by in vivo pathogenicity tests namely, chick embryo and mice inoculation tests. Indirect plate ELISA revealed 41.96% seropositivity for antibodies against listeriolysin O in ewes with abortion or history of abortion and 26.21% in apparently healthy ewes, which after adsorption of sera with Streptolysin O (SLO) reduced to 18.75% and 9.70% in respective groups. On over all basis, out of 215 sheep sera, 74 (34.42%) sera showed positivity for ALLO, which was reduced to 31(14.42%) following adsorption with SLO, indicating the need for sera adsorption for removing the cross-reactivity. The study had significant implications in understanding the epidemiology of listeric infection in migratory flocks.
Keywords | Listeria spp, Migratory sheep, Abortion, LLO, ELISA
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | March 28, 2015; Revised | April 29, 2015; Accepted | April 30, 2015; Published | May 07, 2015
*Correspondence | Shivaramu Keelara, University of Maryland, College Park, USA; Email: shivakeelara@gmail.com
Citation | Keelara S, Malik SSV, Nayakvadi S, Das S, Barbuddhe SB (2015). Isolation and characterization of Listeria spp. from organized and migratory sheep flocks in India. Adv. Anim. Vet. Sci. 3(6): 325-331.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.6.325.331
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Keelara et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Listeriosis caused by pathogenic strains of Listeria monocytogenes and L.ivanovii, is a serious invasive disease characterized by three forms namely; encephalitis, septicaemia and abortion in animals (Low and Donachie, 1997). Listeriosis leads to septicaemia, abortion, stillbirth, perinatal infections, meningitis, gastroenteritis and meningoencephalitis, especially in immunocompromised individuals (Barbudde et al., 2012). The occurrence of listeric infections in the Indian subcontinent has been extensively reviewed by many workers (Malik et al., 2002; Barbudde et al., 2012). In India, we have recorded outbreaks of listeriosis in animal populations (Kumar et al., 2007; Yadav and Roy. 2008; Kaur et al., 2010). Recently, a database of Listeria strains isolated in India from various sources, Indian Listeria Culture Database (ILCD) has been established (Jangam et al., 2010, Barbuddhe et al., 2000; http://www.icargoa.res.in/ilcd). The database provides visualization of geographical source of the strain, its lineage, serotype, source of isolation (animal/human), year of isolation, phenotypic and genotypic characteristics as well as antibiotic sensitivity patterns. Listeric infection in animals is usually acquired by consumption of spoiled silage, in which these bacteria multiply, resulting in herd outbreaks. Though the reliable diagnosis of listeriosis is made by isolation and identification of the pathogen but it is time consuming (Rocourt et al., 1983). Therefore, the diagnosis of pathogenic Listeria spp. and listeric infection can be made on the basis of virulence genes (Notermans et al., 1991b). Listeriolysin-O (LLO), an extracellular 58 kDa haemolysin, is a major virulence factor of L. monocytogenes (Gaillard et al., 1986) and is produced by all the pathogenic strains (Geoffrey et al., 1989). Listeriolysin-O (LLO) expressed by the hly gene serves as an ideal virulence marker as it is produced by only virulent strains of L. monocytogenes.
Detection of virulence factors (LLO and PI-PLC) of pathogenic Listeria spp. (Notermans et al., 1991b) and/or of genes responsible for their expression by PCR assay (Kotlowski et al., 1996) and their correlation with seropositivity in monoclonal or polyclonal antibodies-based ELISA would be an ideal approach for ascertaining the virulence of Listeria isolates from clinical sample. In India, sheep are mainly reared under intensive and semi-intensive rearing systems. Studies on assessment of the extent of listeric infections among sheep population in organized farms/ migratory flocks are lacking. The objective of the study was to investigate the listeric infection in sheep having reproductive disorders and apparently healthy sheep employing various methods.
Materials and methods
Study Population and Collection of Samples
A total of 880 samples comprising blood (n=215), faecal swabs (n=220), vaginal swabs (n=220), placenta (n=10) and serum (n=215) were aseptically collected from ewes (n=220). The animals were apparently healthy (n=103) and clinical cases in ewes with abortion or with a history of abortion (n=117). The samples were collected from one organized sheep farm from Himachal Pradesh (Northern India) as well as three migratory flocks from Karnataka (Southern India) and Himachal Pradesh. All the samples were collected aseptically and transported to the laboratory under chilled conditions and stored at 4oC till processed.
Bacteria
The strains of L. monocytogenes 4b (MTCC 1143), Staphylococcus aureus (MTCC 1144), Rhodococcus equi (MTCC 1135), Streptococcus faecalis (MTCC 439), Bacillus cereus (MTCC 1272), Escherichia coli (MTCC 443), Aeromonas hydrophila (MTCC 646) used in the study were obtained from the Microbial Type Culture Collection and Gene Bank, Institute of Microbial Technology, Chandigarh, India.
The reference strains of Listeria namely, L. monocytogenes 4b (NCTC 11994), L. monocytogenes 1/2a (NCTC 7973), L. monocytogenes 1/2b (NCTC 10887), L. ivanovii (NCTC 11846), Listeria innocua (NCTC 11288), Listeria seeligeri (NCTC 11856), Listeria grayi (NCTC 10812), Listeria welshimeri (NCTC 11857) were kindly provided by Prof. K. L. Morgan, University of Liverpool, U.K. The strains of Salmonella (1117) and Vibrio cholerae (0139) were procured from Division of Veterinary Public Health, Indian Veterinary Research Institute, India.
Isolation of Listeria
Isolation of listeriae from the blood, faecal, vaginal, and placental bits was carried out as per the US Department of Agriculture (USDA) method described by McClain and Lee (1988) after making necessary modifications. Briefly, approx. 5 ml of each of the blood and 25 gm of the placental tissue were inoculated into 50 ml and 225 ml of Pre-enrichment Broth (PEB; Tryptic Soy broth with 0.6% yeast extract; Himedia Laboratories, Mumbai, India). After incubating for 24 h at 30oC, 10 ml of PEB was transferred to 90 ml of University of Vermont-1 (UVM-1) broth (Himedia Laboratories) and incubated overnight at 30oC. Each of the collected swabs (fecal and vaginal) was aseptically inoculated into 10ml of PEB. After incubation, the inoculum was transferred to UVM-1 and incubated overnight at 30oC. The enriched UVM-1 inoculum (0.1ml) was then transferred to UVM-2 broth and again incubated overnight at 30oC. The inoculum enriched in UVM-2 was streaked directly on Dominguez-Rodriguez isolation agar (Dominguez-Rodriguez et al., 1984) and the plates were incubated at 30oC for 48 h. The greenish-yellow glistening, iridescent and pointed colonies of about 0.5mm diameter surrounded by a diffuse black zone of aesculin hydrolysis were suspected to be listeriae. The presumed colonies of Listeria (at least three per plate) were further confirmed.
Confirmation of the Isolates
Morphologically typical colonies were verified by Gram’s staining, catalase reaction, tumbling motility at 20-25oC, methyl red-Voges Proskauer (MR-VP) reactions, CAMP test with S. aureus and R. equi, nitrate reduction, fermentation of sugars (rhamnose, xylose, mannitol and α-methyl-D-mannopyranoside) and hemolysis on 5% sheep blood agar (SBA). The DL-alanine β-napthylamide (DLABN) test was performed as described by McLauchlin (1997) in order to differentiate the listerial isolates.
The biochemically characterized Listeria isolates were plated on ALOA (Himedia Laboratories, Mumbai, India), a chromogenic selective medium for identification of Listeria spp. and their pathogenic potential as described by Ottaviani et al. (1997). Each Listeria isolate was spot inoculated separately on ALOA plates and incubated at 37oC and then observed for typical blue-green colonies with clearly defined opaque halo up to 5 days.
The pathogenicity testing of the Listeria isolates was done by mice inoculation test (Menudier et al., 1991) and chick embryo inoculation (Notermans et al., 1991b) and permission from institute animal ethical committee (IAEC) was obtained before testing.
Phosphatidylinositol-specific Phospholipase C (PI-PLC) Assay
All the biochemically characterized Listeria isolates were assayed for PI-PLC activity as per the method of Notermans et al. (1991a) with certain modifications. In brief, the Listeria isolates were grown overnight onto sheep blood agar (SBA) plates at 37oC. The growth of each Listeria isolate harvested from the SBA plate was spot inoculated separately on tryptone soya yeast extract (TSYE, Himedia Labs, Mumbai, India) agar plates supplemented with 2.5 mM CaCl2 and 40 mM MgSO4 in a manner to get a clear visible bacterial growth of approximately 2 mm diameter following an incubation at 37oC for 24 to 48 h.
The L-α-phosphatidylinositol (PI) solution was prepared by dissolving PI substrate (Sigma, USA) in 20 mM Tris–HCl (Sigma) buffer pH 7.0 at 20 mg/ml and subjecting the resultant turbid solution to ultrasonication (5 cycles of 60 s each with 30 s cooling intervals) in an ultrasonicator (Sanyo, U.K.). Subsequently, the agarose solution (1.4%) prepared in 20 mM Tris– HCl buffer, pH 7.0 was cooled to 55oC and added with 100 μg/ ml of chloramphenicol to prevent the growth of contaminants. Finally, an overlay suspension was prepared by mixing the PI solution pre-warmed to 55oC in a water bath with an equal volume of agarose solution held at 55oC just before the use and overlaid at a rate of 4 ml per petri dish (9cm diameter) onto the previously seeded colonies of Listeria isolates on TSYE. The plates were then incubated at 37oC and observed daily for turbid halos around colonies up to 5 days.
Multiplex Polymerase Chain Reaction (mPCR) Assay for the Detection of Virulence Genes of Listeria spp.
All the Listeria isolates were screened by multiplex polymerase chain reaction (mPCR) as per the protocol and primer sequences used by Rawool et al. (2007) for the detection of virulence associated genes viz., haemolysin (hlyA), Phosphatidylinositol-specific phospholipase C (plcA), ActA protein (actA), a surface protein, p60 (iap) and positive regulatory factor A (prfA). Briefly, the mPCR protocol was standardized employing the standard pathogenic strains of L. monocytogenes 4b (MTCC 1143, NCTC 11994), L. monocytogenes ½a (NCTC 7973), L. monocytogenes ½b (NCTC 10887) and L. ivanovii (NCTC 11846). Subsequently, the test isolates were screened by the standardized mPCR for the detection of aforesaid virulence associated genes of Listeria spp.
50 μl PCR reaction mix included 5.0 μl of 10× PCR buffer (100 mM Tris–HCl buffer, pH 8.3 containing 500 mM KCl, 15 mM MgCl2 and 0.01% gelatin), 0.2 mM dNTP mix (Sigma, USA), 2 mM MgCl2 and 10 μM of a primer set containing forward and reverse primers (a final concentration of 0.1 μM of each primer), 1 U of Taq DNA polymerase (Sigma, USA), 5 μl of cell lysate and sterilized milliQ water to make up the reaction volume. The DNA amplification reaction was performed in a Master Cycler Gradient Thermocycler (Eppendorf, Germany) with a preheated lid. The cycling conditions for PCR included an initial denaturation of DNA at 95°C for 2 min followed by 35 cycles each of 15 s denaturation at 95°C, 30 s annealing at 60°C and 1 min and 30 s extension at 72°C, followed by a final extension of 10 min at 72°C and held at 4°C. All the five sets of primers for virulence-associated genes were amplified under similar PCR conditions and amplification cycles. The resultant PCR products were further analyzed by agarose gel electrophoresis (1.5%; low melting temperature agarose L), stained with ethidium bromide (0.5μg/ml) and visualized by a UV transilluminator (UVP Gel Seq Software, England).
ELISA
Indirect plate ELISA was standardized to detect antibodies in the sera of spontaneous cases of listeric infection in aborted ewes and also in apparently healthy ewes. Listeriolysin-O (LLO) was prepared and purified from the cell free supernatant (CFA) by ion exchange chromatography in accordance with the method of Lhopital et al. (1993). The purity of the LLO was checked by SDS-PAGE, which showed a homogeneous 58.0 kDa protein. The fractions having LLO were pooled and the protein content was estimated, and finally stored at -20oC until used.
The indirect plate ELISA was performed as per the methods of Low and Donachie (1991). The ELISA was standardized by checker board analysis. Briefly, purified LLO was used in at the optimal concentration 40 ng/well (100µl/well) for coating the microtitre plates (Nunc, Denmark). The plates were covered and incubated at 37oC for 2 h before washing five times with phosphate buffer saline (PBS), pH 7.2 plus 0.05% Tween 80 (PBS-T). Each of the test serum collected from aborted, with history of abortion and apparently healthy ewes was diluted 1:200 in PBS and added (100µl/well) in duplicate to the plates. The sealed plates were incubated at 37oC for 90 min and again washed as before. Subsequently, rabbit anti-sheep IgG HRPO conjugate (Bethyl, India) was diluted to 1:10000 in PBS were added (100 µl/well) to the plates. The plates were again incubated at 37oC for 90 min and washed as described earlier. Finally, ortho-phenylenediamine dihydrochloride (OPD) solution (1 mg/ml) in citrate buffer with 12 µl/100 ml of hydrogen peroxide was added (100µl/well) as a substrate and after 15 min, the reaction was read by ELISA plate reader (Anthos Labtek) at 492 nm. A serum sample at a dilution of 1:200 with a positive to negative (P/N) ratio of more than 2 was considered positive for listeriosis in standardized ELISA.
The detection of specific ALLO which is often present at low titer in ewe sera requires prior adsorption of anti-SLO, as Streptolysin-O (SLO) and Listeriolysin-O (LLO) are antigenically related. The adsorption of anti-SLO in the sera was done as per the method described by Berche et al. (1990) with certain modifications. The purified SLO (Sigma-Aldrich, USA) was coated onto 96-well flat bottom polysorp plates (Nunc, Denmark) at a concentration 60 µg/ml, which was added to wells at the rate of 200µl/well and then incubated for 1 h at 25oC. Each ewe serum (0.5ml), diluted 1/100 and 1/200 in PBS-T was then added to the wells and then incubated for 1 h at 25oC. The SLO-adsorbed ewe sera were then used in the indirect plate ELISA to screen the ewe sera for ALLO as per the method described earlier.
Results
Isolation and Identification of Pathogens
Different Listeria spp. were isolated from 23 of 665 (225 each of fecal and vaginal swab, 10 placenta and 215 of blood) samples collected from 220 ewes. Out of these 15 isolates were confirmed as L. monocytogenes, two isolates as L. ivanovii and the remaining were Listeria spp. No Listeria spp. could be isolated from blood samples. The overall occurrence of listeric infection was 3.4%.
Pathogenicity Testing
Out of these 23 isolates, four haemolytic isolates (L. ivanovii-2 and L. monocytogenes-2) were found to be pathogenic by in vitro pathogenicity tests namely, haemolysis on sheep blood agar (SBA); Christie, Atkins, Munch-Petersen (CAMP) test, phosphatidylinositol-specific phospholipase C (PI-PLC) activity on ALOA medium and in PI-PLC assay as well as by in vivo pathogenicity tests namely, chick embryo and mice inoculation tests (Table 1). All other Listeria spp. isolates were non-pathogenic.
Multiplex Polymerase Chain Reaction Assay
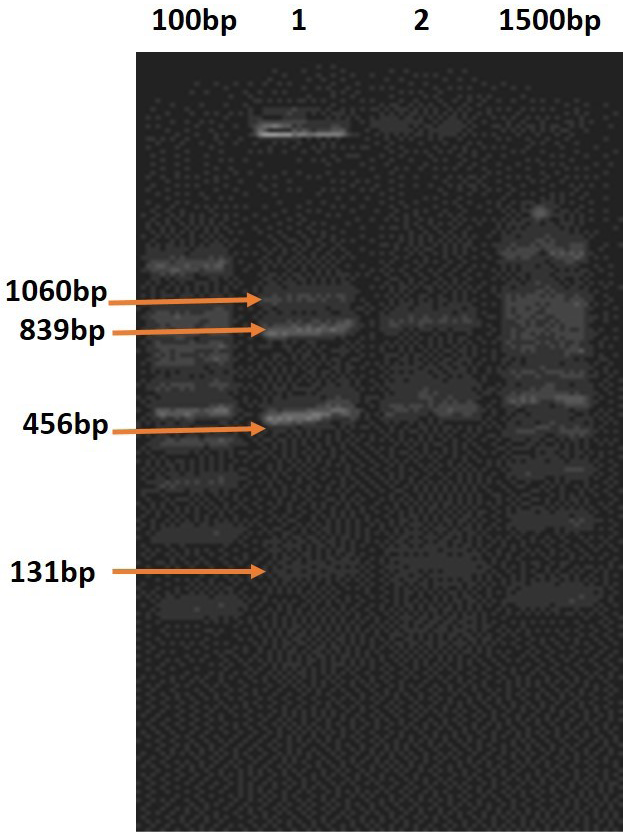
In multiplex PCR, both the primer sets employed i.e., one comprising primers for plcA, actA, hlyA and iap, and the other for prfA, actA, hlyA and iap, amplified the DNA fragments of the expected sizes. Out of four haemolytic Listeria isolates ( L.monocytogenes -2 and L.ivanovii-2) The DNA of L. ivanovii isolates recovered from ewes with history of abortion revealed amplified products corresponding to all the five virulence-associated genes namely, plcA, prfA, actA, hlyA and iap in mPCR employing the above indicated sets of primers in 2 separate reactions (Figure 1).
On DNA analysis by the mPCR, out of the two hemolytic, ALOA-positive and PI-PLC-positive L. monocytogenes isolates recovered from abortion case and apparently healthy ewe, respectively, the former revealed the amplified products of all the five virulence-associated genes while the later did not show plcA gene (Table 1).
ELISA
The antibodies against LLO (ALLO) were detected in 47 out of 112 (41.96%) serum samples from ewes with a history of abortion and 27 out of 103 (26.21%) apparently healthy cases. However, after adsorption with SLO and retesting by plate ELISA, the positivity for ALLO reduced to 21 (18.75%) of clinical cases and 10 (9.7%) of apparently healthy cases (Table 2).
Table 1: Pathogenicity and PCR profiles of L. monocytogenes and L.ivanovii isolates from healthy and clinical cases in ewes
|
Isolate No. |
Source of the isolate |
Pathogenicity profile |
PCR profile of virulence-associated genes |
||||||||||
|
In vitro tests |
In vivo tests |
||||||||||||
|
Hemolysis on SBA |
ALOA |
CAMP with S/R |
PI-PLC assay |
Mice lethality |
Chick embryo lethality |
plcA |
prfA |
actA |
hlyA |
iap |
|||
|
Col |
Halo |
||||||||||||
|
Isolate 1 |
Abortion (Place- nta) |
++ |
BG |
+++ |
+S |
++ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Isolate 2 |
History of abortion (fecal swab) |
++ |
BG |
++ |
+R |
++ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Isolate 3 |
History of abortion (Vaginal swab) |
++ |
BG |
++ |
+R |
++ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Isolate 4 |
Healthy (fecal swab) |
++ |
BG |
++ |
+S |
++ |
+ |
+ |
- |
+ |
+ |
+ |
+ |
PI-PLC = Phosphatidylinositol-specific phospholipase C; PCR = Polymerase chain reaction; SBA = Sheep blood agar; S/R = Staphylococcus aureus / Rhodococcus equi; ALOA = Agar Listeria according to Ottaviani and Agosti; ALLO = Antibodies against Listeriolysin-O; CAMP = Christie, Atkins, Munch-Petersen; BG = Blue green; L.m. = L. monocytogenes; L.i = L. ivanovii
Table 2: Seropositivity of ewes against listeriolysin-O (LLO) with clinical cases and apparently healthy ewes
|
Sl No |
Clinical status |
No of samples collected |
No of samples Positive |
% seropositivity |
Adsorption with SLO |
|
1 |
Abortion |
112 |
47 |
41.96 |
21 (18.75%) |
|
2 |
Healthy |
103 |
27 |
26.21 |
10 (9.7%) |
|
3 |
Total |
215 |
74 |
34.42 |
31 (14.42%) |
Discussion
The genus-Listeria has two pathogenic species namely, Listeria monocytogenes and L. ivanovii. Of these, L. monocytogenes is a well-known cause of abortion, encephalitis and septicaemia in animals and human beings (Rocourt and Seeliger, 1985; McLauchlin, 1987; Kumar et al., 2007). The other pathogenic spp, L. ivanovii was isolated from cases of abortion, still birth and neonatal septicemia in sheep and cattle (Jose et al., 2001). There are many pathogenic spp of Listeria known to be excreted in faeces and vaginal secretions which can act as a source of infection in the environment. The listeric infection in animals is mainly transmitted by consumption of spoiled silage, aborted materials and vaginal secretions in which these bacteria multiply readily, resulting in outbreaks, especially in the sheep (Low and Donachie, 1997; Kumar et al., 2007). In our study, the pathogenic and non-pathogenic spp. of Listeria were recovered from both organized and migratory flocks from different regions. Isolation of pathogenic strains such as L. ivanovii and L. monocytogenes from aborted ewes in this study showed that, these two isolates are potent pathogens of reproductive disorders in sheep. The migratory flocks had abortion and a history abortion. It is difficult to comment on the route of infection among these sheep but infection might have transmitted from ingestion of infected material or soil.
Pathogenic isolates of L. ivanovii were recovered from 2 cases with a history of abortion while pathogenic isolate of L. monocytogenes was recovered from one case of spontaneous abortion. These results are in agreement of other published work wherein pathogenic L. ivanovii has been found to be associated with 4.6% cases of abortion in ewes (Nigam et al., 1999) whereas L. monocytogenes has been recovered from 9.1% (Nigam et al., 1999) and 15.7% (Sharma et al.,1996) cases of abortion in ewes from migratory flocks from Himachal Pradesh. The results of present investigation are also in line with earlier reports. L. ivanovii has been implicated as the most frequent cause of abortions in case of sheep (McLauchlin, 1987), with abortion rates ranging from 1.65% (Alexander et al.,1992) to 12% (Sergeant et al.,1991). One pathogenic L. monocytogenes isolate was recovered from apparently healthy ewe. L. monocytogenes has been found to translocate throughout the digestive tract in asymptomatic sheep (Zundel et al., 2006).
A number of factors are involved in the manifestation of virulence of L. monocytogenes (Portnoy et al. 1992; Vazquez-Boland et al., 2001). It has been clearly demonstrated that L. monocytogenes phospholipases are essential determinants of pathogenicity (Smith et al., 1995). In the present investigation, even though expression of haemolytic activity by all 23 isolates only four isolates were found to be pathogenic in all the assays and possessed all the five virulence genes except one L. monocytogenes isolate lacking of the plcA gene. Truncated form or mutation in the plcA gene might have contributed to this observation. Those isolates which are PI-PLC negative turned out negative in mice and chick embryo assays.
In the present study, a relatively high (41.96%) seropositivity was observed against LLO in ewes with abortion or a history of abortion. In case of apparently healthy ewes, relatively less seropositivity (26.21%) was observed. The observation is comparable to earlier reports as 33% documented by Barbuddhe et al. (2000). However, on testing of the sera adsorbed by SLO, the per cent seropositivity decreased. Out of 47 sera from ewes with abortion or history of abortion showing seropositivity, 21 were seropositive after adsorption with SLO. Ten sera from apparently healthy ewes were seropositive after adsorption with SLO. The reduction of seropositivity is in agreement with the earlier published report on the reduction of the titer from 100 before adsorption to less than 12.5 after adsorption of the cross-reacting whole human sera with S. aureus (Larsen and Jones, 1972). Kaur et al. (2006) reported reduction in seropositivity from 48% to 16% after adsorption with SLO. Thus, from the present study it is evident that adsorption of the sera eliminates cross-reactions in serological tests.
The recovery of pathogenic isolates of L. monocytogenes and L. ivanovii from clinical cases and apparently healthy ewes indicated association of this pathogen with such cases. The study highlights the role of listeric infection in causing reproductive disorders in organized was well as migratory sheep. This study provides significant implications for understanding the epidemiology of listeric infection and is required for control of infection.
References