Advances in Animal and Veterinary Sciences
Research Article
Molecular Identification of Aeromonas hydrophila Isolate with Sensitivity and Resistance to Antibiotics for its Different Strains
Md. Zobayer Rahman1, Arman Hossain1, Md. Moshiur Rahman1, Shamima Nasren2, Md. Abdullah Al Mamun1*, Sarker Mohammed Ibrahim Khalil1, M. M. Mahbub Alam1
1Department of Fish Health Management, Faculty of Fisheries, Sylhet Agricultural University, Sylhet- 3100, Bangladesh; 2Department of Fish Biology and Genetics, Faculty of Fisheries Sylhet Agricultural University, Sylhet- 3100, Bangladesh.
Abstract | Bacteria plays a vital role in the incidence of fish diseases both in fresh and saline water, thus antibiotics are used for controlling these diseases. For effective treatment, the causative agents of the diseases need to be identified properly. The study was carried out to identify one Aeromonas hydrophila isolated from walking catfish Clarias batrachus and to determine the sensitivity and resistance of five different strains of A. hydrophila to 15 commercially available antibiotics. Aeromonas hydrophila isolated from the diseased walking catfish (C. batrachus) was initially confirmed, using Rimler- Shotts (RS) selective medium for the isolation of A. hydrophila characterized by yellow round colonies. The obtained 16S rRNA sequence of A. hydrophila matched with 887236 – 888648 bps (Identity- 99.72%) of the 16S rRNA gene of A. hydrophila strain B11 chromosome complete genome (GB Accession number CP053859.1). The identity of the sequence with the A. hydrophila sequences in NCBI confirmed that the bacteria isolated from C. batrachus was A. hydrophila and responsible for the disease Motile Aeromonas Septicemia (MAS) or Dropsy. Five strains of A. hydrophila used in this study were completely resistant to ampicillin sulbactam (20 µg) and oxacillin (10 µg). Amoxicillin (30 µg) was moderately resistant due its variant response over antibiotics. The rest of the antibiotics were intermediate and sensitive to A. hydrophila. Ciprofloxacin (5 μg) and Levofloxacin (5 μg) can be used to treat bacterial infections caused by one of the A. hydrophila strains. The information of the present study will be helpful to identify A. hydrophila and the antibiotics to be used to control the bacterial agent in aquaculture.
Keywords | Antibiotics, Sensitivity, 16S rRNA gene, Aeromonas hydrophila, Clarias batrachus
Received | June 04, 2021; Accepted | August 20, 2021; Published | October 15, 2021
*Correspondence | Md. Abdullah Al Mamun, Department of Fish Health Management, Faculty of Fisheries, Sylhet Agricultural University, Sylhet- 3100, Bangladesh; Email: maamamun.fhm@sau.ac.bd
Citation | Rahman MZ, Hossain A, Rahman MM, Nasren S, Mamun MMA, Khalil SMI, Alam MMM (2021). Molecular identification of Aeromonas hydrophila isolate with sensitivity and resistance to antibiotics for its different strains. Adv. Anim. Vet. Sci. 9(12): 2062-2068.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.12.2062.2068
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Rahman et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Among the infectious diseases in aquatic organisms, bacterial diseases are the most common challenge causing much mortality. Wide ranges of fish bacteria have the potential to cause diseases. Among them the genus Aeromonas is one of the main pathogenic bacterial genera, which are extensively inhabit the aquatic environment. The facultative, anaerobic, oxidase-positive and gram-negative genus Aeromonas bacteria generally found in aquatic environment (Igbinosa et al., 2012) and notable as a significant disease producing agent of fish and other cold and warm-blooded organisms (Krishnakumar et al., 2009). Aeromonas hydrophila is the opportunistic pathogen commonly inhabits the digestive region of fish (Yildiz et al., 2005). Many diseases including Motile Aeromonad Infection (MAI) were caused by this aetiological agent in wide variety of cultured fishes (Mirand and Zemelman, 2002; Michael et al., 2003). Motile aeromonas infection is one of the main bacterial diseases for commercial fish farming.
The most common method of monitoring bacterial disease is the use of antimicrobial drug, but proper identification of strains of bacteria or infectious agent are necessary. Most of the cases, researchers attempted to identify bacteria using specific culture media and biochemical tests, but it is difficult to identify bacteria strains and serotypes using these traditional methods (Miñana‐Galbis et al., 2002; Frans et al., 2008; Citarasu et al., 2011; Erdem and Kar, 2011; Samal et al., 20014; Goni et al., 2020; Parven et al., 2020). In contrast, bacterial strains can be accurately identified using molecular techniques of which PCR- Polymerase Chain Reaction and gene sequencing were found effective (Frans et al., 2008; Balsalobre et al., 2009; Trakhna et al., 2009; Hu et al., 2012; Oliveira et al., 2012; Mansour et al., 2019).
Substantial quantity of antibiotics has been used with supplementary feed for the prevention and control of bacterial diseases in aquaculture worldwide (Sapkota et al., 2008). Aquatic bacteria become resistant to antibiotics due to indiscriminate use of antibiotics to treat bacterial diseases (Vivekanandhan et al., 2002). Resistance to antimicrobial agents is common and occurs in several bacterial species (Chopra and Roberts, 2001). Possibly, the strains which are resistant have effect on treatment of aquatic animal diseases with its environment (Smith et al., 2003). In developing countries, antibiotics are widely used in aquaculture and this is more intricate where the uses of antibiotics are not controlled and regulated. Many researchers from different parts of the World isolated A. hydrophila which were multi-resistant and found sensitive to cephalosporins 2nd and 3rd generations, trimethoprim-sulfamethoxazole, quinolones, aminoglycosides, tetracycline and chloramphenicol, but resistant to ampicillin and penicillin (Emekdas et al., 2006). Limited data were found on the antimicrobial resistance of bacteria in aquatic organisms including fish as well as the aquaculture environment. Therefore, this research was carried out to detect A. hydrophila isolate more effectively using molecular technique and then to determine the sensitivity and resistance of its different strains against different antibiotics.
Results and Discussion
Identification of Aeromonas hydrophila
Aeromonas hydrophila, which was responsible for Motile Aeromonas Seticemia (MAS) or Dropsy and isolated from the body cavity of a diseased walking catfish (C. batrachus) was initially confirmed, using Rimler- Shotts (RS) selective medium for the isolation of A. hydrophila (Himedia, Mumbai). The RS medium culture was characterized by yellow round colonies (Supplementary Figure 1). The documentation following gel electrophoresis confirmed that the PCR for A. hydrophila 16S rRNA gene produced about 1,450 bp length PCR product. The sequencing of A. hydrophila from pure culture produced 1,419 bps length sequence with a molecular weight of 430304 Daltons (single strand) and nucleotide composition of A – 24.45%, C – 23.40%, G – 32.56% and T – 19.59%. The obtained 16S rRNA sequence (GenBank Accession number: MZ046725) matched with 887236 – 888648 bps (Query cover-99% and Identity- 99.72%) of the 16S rRNA gene of A. hydrophila strain B11 chromosome complete genome (GenBank Accession number CP053859.1). The sequence also matched with A. hydrophila sequences, having GenBank accession numbers: AM992197.3 (Query cover- 99%, Identity- 99.72%), LC420120.1 (Query cover- 99%, Identity- 99.58%), JQ040106.1 (Query cover- 98%, Identity- 99.57%) and AB473028.1 (Query cover- 99%, Identity- 99.50%). Thus, the identity (Maximum 99.72%) of the sequence with the A. hydrophila sequences in NCBI confirmed that the bacteria isolated from C. batrachus was A. hydrophila.
Figure 1: Map of Bangladesh showing the sapling site of walking catfish (Clarias batrachus). Letter: SB- Singari Beel.
Sensitivity and resistance to antibiotics
The zone of inhibition developed by A. hydrophila strains presented various responses for different antibiotics (Table 1). Different A. hydrophila strains used here were completely resistant to ampicillin sulbactam (20 μg) and oxacillin (10 μg). Amoxicillin (30 μg) showed no zone of inhibition in the petri dish (0 cm) in (MTCC 1739) starin, A. hydrophila from magur (Clarias batrachus) and zebra (Danio rerio). In the petri dish of ATCC 36562 strain and A. hydrophila from koi carp (Cyprinus rubrofuscus) developed a tiny zone of inhibition hence considered as resistant (Figure 2; Supplementary Figure 2).
Aeromonas hydrophila were recorded highly sensitive (> 3cm) towards levofloxacin (5 μg) and ciprofloxacin (5 μg) (Table 1; Supplementary Figure 2). Ampicillin Sulbactam (20 μg) and Oxacillin (10 μg) showed full resistance against A. hydrophila strains isolated from various sources (Table 2; Figure 2; Supplementary Figure 2). On the other hand, Ciprofloxacin (5 μg) and Levofloxacin (5 μg) have shown their full sensitivity (100%) to all A. hydrophila strains/isolates. Amoxicillin (30 μg) scored as moderate resistant considering its length of inhibition. Aeromonas hydrophila showed moderate sensitivity to tetracycline (30 μg), azithromycin (15 μg), ceftriaxone (30 μg), doxycycline hydrochloride (30 μg), streptomycin (10 μg), oxytetracycline (30 μg), gentamycin (10 μg), erythromycin (15 μg) and novobiocin (30 μg) (Tables 1, 2; Figure 2; Supplementary Figure 2).
Table 1: Sensitivity of five Aeromonas hydrophila strains and isolates to selected antibiotics.
| S. No. | Short form | Antibiotics name |
A. hydrophila strains and isolates |
||||
| ATCC (cm) | MTCC (cm) | Koi (cm) | Zebra (cm) | Magur (cm) | |||
| 01. | CIP | Ciprofloxacin (5 μg) | 2.9(+++) | 3.1(+++) | 3.0(+++) | 3.9(+++) | 2.3(+++) |
| 02. | A/S | Ampicillin Sulbactam (20 μg) | 0(-) | 0(-) | 0(-) | 0(-) | 0(-) |
| 03. | TE | Tetracycline (30 μg) | 1.3(++) | 0.7(+) | 1.1(++) | 0.9(+) | 1.6(++) |
| 04. | AT | Azithromycin (15 μg) | 1.7(++) | 3.5(+++) | 1.9(++) | 1.5(++) | 2.8(+++) |
| 05. | CTR | Ceftriaxone (30 μg) | 1.1(++) | 0.7(+) | 0.9(+) | 2.5(+++) | 0(-) |
| 06. | LE | Levofloxacin (5 μg) | 3.1(+++) | 3.2(+++) | 3.1(+++) | 3.9(+++) | 2.7(+++) |
| 07. | DO | Doxycycline hydrochloride (30 μg) | 2.7(+++) | 2.0(++) | 1.6(++) | 2.2(+++) | 2.0(++) |
| 08. | COT | Co-Trimoxazole (25 μg) | 1.0(++) | 1.8(++) | 1.6(++) | 2.9(+++) | 0(-) |
| 09. | S | Streptomycin (10 μg) | 2.9(+++) | 2.9(+++) | 2.9(+++) | 2.5(+++) | 0.8(+) |
| 10. | O | Oxytetracycline (30 μg) | 2.5(+++) | 1.6(++) | 1.5(++) | 2.1(+++) | 1.7(++) |
| 11. | GEN | Gentamycin (10 μg) | 2.6(+++) | 2.6(+++) | 2.6(+++) | 2.3(+++) | 1.7(++) |
| 12. | OX | Oxacillin (10 μg) | 0(-) | 0(-) | 0(-) | 0(-) | 0(-) |
| 13. | E | Erythromycin (15 μg) | 1.1(++) | 3.2(+++) | 2.9(+++) | 1.7(++) | 2.6(+++) |
| 14. | NV | Novobiocin (30 μg) | 2.5(+++) | 2.3(+++) | 2.3(+++) | 0.9(+) | 1.2(++) |
| 15. | AMX | *Amoxicillin (30 μg) | 0.5(+) | 0(-) | 0.8(+) | 0(-) | 0(-) |
(-): No inhibition in media considered as Resistant; (+): Inhibitory zone (0-1) cm considering as intermediate; (++): Inhibitory zone between (1-2) cm as moderate; (+++): Inhibitory zone equal (2 to >3) cm as Sensitive.
Table 2: Antibiogram profile percentages (%) of isolated colonies (n=5).
| S. No. | Short form | Antibiotics name | Sensitive | Intermediate | Resistant |
| 01. | CIP | Ciprofloxacin (5 μg) | 5(100%) | 0 | 0 |
| 02. | A/S | Ampicillin Sulbactam (20 μg) | 0 | 0 | 5(100%) |
| 03. | TE | Tetracycline (30 μg) | 0 | 3(60%) | 2(40%) |
| 04. | AT | Azithromycin (15 μg) | 2(40%) | 3(60%) | 0 |
| 05. | CTR | Ceftriaxone (30 μg) | 1(20%) | 3(60%) | 1(20%) |
| 06. | LE | Levofloxacin (5 μg) | 5(100%) | 0 | 0 |
| 07. | DO | Doxycycline hydrochloride (30 μg) | 2(40%) | 3(60%) | 0 |
| 08. | COT | Co-Trimoxazole (25 μg) | 1(20%) | 3(60%) | 1(20%) |
| 09. | S | Streptomycin (10 μg) | 4(80%) | 0 | 1(20%) |
| 10. | O | Oxytetracycline (30 μg) | 3(60%) | 2(40%) | 0 |
| 11. | GEN | Gentamycin (10 μg) | 4(80%) | 1(20%) | 0 |
| 12. | OX | Oxacillin (10 μg) | 0 | 0 | 5(100%) |
| 13. | E | Erythromycin (15 μg) | 3(60%) | 2(40%) | 0 |
| 14. | NV | Novobiocin (30 μg) | 3(60%) | 1(20%) | 1(20%) |
| 15. | AMX | Amoxicillin (30 μg) | 0 | 0 | 5(100%) |
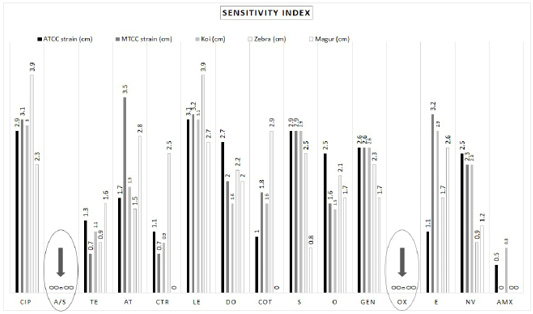
Figure 2: Bar Diagram representing total 15 antibiotics and their length of inhibition. Letters: CIP, Ciprofloxacin; A/S, Ampicillin/Sulbactam; TE, Tetracycline; AT, Azithromycin; CTR, Ceftriaxone; LE, Levofloxacin; DO, Doxycycline; COT, Co-Trimoxazole; S, Streptomycin; O, Oxytetracycline; GEN, Gentamycin; OX, Oxacillin; E, Erythromycin; NV, Novobiocin; AMX, Amoxycillin.
Identification of Aeromonas hydrophila
Unlike the present study, identification of A. hydrophila, using Rimler-Shotts (RS) selective medium and subsequently utilizing molecular methods, is very common in the World. Samal et al. (2014) identified A. hydrophila strain based on small, round, smooth, convex and translucent, yellow colonies on RS medium and 34 different sugar fermentation tests. Rashid et al. (2013) identified A. hydrophila based on specific morphology, physiological and biochemical characteristics. Aboyadak et al. (2015) confirmed 12 strains of A. hydrophila utilizing A. hydrophila specific 16S rRNA gene primer. Aeromonas hydrophila sequences amplified using 27F and 1492R 16S rRNA specific primers which showed 99% similarity with standard A. hydrophila sequence in NCBI (Yazdanpanah-Goharrizi et al., 2020).
Sensitivity and resistance to antibiotics
Different studies tested different antibiotics on A. hydrophila to know its sensitivity towards the antibiotics and found a wide range of sensitivities. Aeromonas hydrophila isolates were found resistant to ampicillin, amoxicillin, amoxicillin-clavulanic acid, oxytetracycline, and streptomycin (Turutoglu et al., 2005).
Antibiotics resistance to A. hydrophila is a fixed problem in the global fish farming (Harikrishnan and Balasundaram 2005). Radu et al. (2003) noted multiple antibiotic resistance of different Aeromonas spp. to different antibiotics including ampicillin and tetracycline. Our results corroborated with the findings of Belém-Costa et al. (2006) where they found the antimicrobial activity in bacterial isolates from tilapia were resistant to amoxicillin, ampicillin, lincomycin, novobiocin, oxacillin, penicillin, and trimethoprim + sulfamethoxazole as well as from pacu.
Aeromonas hydrophila strains isolated from various sources was found resistant to ampicillin sulbactam (20 μg) and oxacillin (10 μg) showed full resistant as expected, because A. hydrophila naturally resistant to ampicillin. Aeromonas hydrophila strain resistant to tetracycline were very common (Kampfer et al., 1999). Aeromonas hydrophila isolates from 15 fishes exhibited resistant to ampicillin and colistin antibiotics, and moderate sensitivity to co-trimoxazole (41.8%) and oxytetracycline (50%) (Kaskhedikar and Chhabra, 2010). Many antibiotics including erythromycin, streptomycin and carbenicillin were found to be resistant against different Aeromonas spp. (Radu et al., 2003). Earlier studies showed that Aeromonas spp. isolated from diseased fishes were 100% sensitive to ciprofloxacin (5μg) but resistant to ampicillin (Hamom et al., 2020; Parven et al., 2020). The sensitivity of 23 different antibiotics tested on another species under the genus Aeromonas (i.e., A. salmonicida) which was also pathogenic to fishes, using disc diffusion method resulted all the strains susceptible but only ampicillin and venomycin were resistant (Bektas et al., 2007). Our earlier studies exhibited intermediate sensitive to azithromycin (15μg), tetracycline (30μg) and streptomycin, is also evident in the present study (Goni et al., 2020; Hamom et al., 2020; Parven et al., 2020). Now a days, A. hydrophila controlling becomes a fixed problem in the modern intensive aquaculture firms, hence appropriate antibiotics prerequisite for the sustainable fish health management.
To conclude, bacterial diseases cause significant loss both in fresh and saline water aquaculture production. Antibiotics and other chemotherapeutic drugs are used indiscriminately for the management of diseases without proper identification of causing agents and effective drugs. The present study showed a process to identify pathogenic bacteria with its sensitivity to antibiotics. Aeromonas hydrophila was initially confirmed, using Rimler-Shott’s (RS) selective medium and finally by sequencing the 16S rRNA sequence which showed 99.72% identity with the A. hydrophila complete genome (GenBank Accession number CP053859.1). The five strains of A. hydrophila were completely resistant to ampicillin sulbactam (20 μg) and oxacillin (10 μg). Aeromonas hydrophila was most sensitive to Ciprofloxacin (5 μg) and Levofloxacin (5 μg) followed by Gentamycin (10 μg), Streptomycin (10 μg) and Erythromycin (15 μg). The patterns of resistant to antibiotics should be frequently examined to guess the initiation and prevalent of multiple antibiotic resistance. Definite task is necessary to possess the new resistance from further emerging and dissemination. Otherwise, the presence of antimicrobial resistant in A. hydrophila poses threats to aquatic biota and public health. The information of the present study will helpful to identify A. hydrophila and the antibiotics to be used to control the bacterial agent in aquaculture as well as ornamental fishery. As bacteria are evolving with the time, environmental changes and condition of the hosts, new antibiotics should be developed to treat the future virulent strains of A. hydrophila.
MATERIALS AND METHODS
Sample collection
A diseased walking catfish, locally known as magur, Clarias batrachus was collected from Singari Beel (24°52’07” N; 91°56’52”) of Sylhet, Bangladesh (Figure 1) and transported in live condition to Fish Lab for bacteria sample collection.
Isolation of Aeromonas hydrophila
Two strains (ATCC 36562 and MTCC 1739) and two isolates of A. hydrophila from koi carp Cyprinus rubrofuscus and zebra fish Danio rerio respectively were collected at Department of Aquatic Animal Health Management, College of Fisheries, Mangalore, Karnataka, India by one of the co-author of this study, but their sensitivity and resistance to antibiotics were not tested. Besides, inclusion of the mentioned strains and isolates facilitated the comparison with the newly isolated A. hydrophil (Fifth isolate) from diseased magur (Clarias batrachus). The fifth A. hydrophil isolate were isolated from diseased magur (Clarias batrachus) following a standard protocol in the Central Laboratory, Faculty of Fisheries, Sylhet Agricultural University (Mamun et al., 2019). Briefly virulent A. hydrophila separated from the body cavity of an infected walking catfish (C. batrachus) was injected in climbing perch (Anabas testudineus) and re-isolated, using Rimler- Shotts (RS) selective medium for the isolation of A. hydrophila (Himedia, Mumbai). Kidney and gill surface of the diseased fish were swabbed and streaked on the RS agar plate. After one day of incubation at 37oC, yellow colonies were grown on the RS medium. The pure yellow colonies from RS agar plates were randomly picked, and stored as streaks in Brain Heart Infusion (BHI) agar (Himedia, Mumbai) slants.
DNA extraction of Aeromonas hydrophila isolated from Clarias batrachus
DNA of A. hydrophila from pure culture was extracted using Maxwell Blood DNA Kit, Model: AS1010, Origin: Promega, USA.
PCR amplification and sequencing of Aeromonas hydrophila
PCR of 16S rRNA was performed in a final volume of 20 µL, utilizing Hot Start Green Master Mix composed of dNTPs, Buffer, MgCl2, and Taq Polymerase (Promega, USA), using two primers i.e., 27F (5՜- AGA GTT TGA TCM TGG CTC AG-3՜) and 1492R (5՜- CGG TTA CCT TGT TAC GAC TT-3՜). The amplification protocol for 16S rRNA gene included three minutes denaturation at a temperature of 95°C, then 30 s denaturation at a temperature of 95°C (35 cycles), 30 s annealing at a temperature of 48°C (35 cycles) and 90 s extension at a temperature of 72°C (35 cycles), and finally 5 min extension at a temperature of 72°C. The product of PCR amplification was tested on agar gel using Gel electrophoresis (Origin: Promega, USA), 100 bp DNA Ladder (Promega, USA), 1kb DNA Ladder (Promega, USA), Diamond™ Nucleic Acid Dye (Promega, USA) and TAE Buffer (Promega, USA).
After purification of the PCR product, the sequencing PCR was performed, using Big Dye Terminator kit and either the forward or the reverse primers. Then the DNA template was precipitated using ethanol. Finally, sequencing was performed in a 3500xL Genetic Analyzer (AB).
Sensitivity and resistance to antibiotics
All strains and isolates of A. hydrophila were inoculated in broth nutrient media and incubated at 37 oC for overnight later, streaked into Mueller-Hinton agar by using sterile cotton swab. Disc diffusion method for antibiotic susceptibility was conducted as described by Guz and Kozinska (Guz and Kozinska, 2004). The research work were done prior to the permission of the Animal Ethics Committee, Sylhet Agricultural University.
A total of 15 antibiotic impregnated discs with their concentrations: Ciprofloxacin (5 μg), ampicillin sulbactam (20 μg), tetracycline (30 μg), azithromycin (15 μg), ceftriaxone (30 μg), levofloxacin (5 μg), doxycycline hydrochloride (30 μg), co-trimoxazole (25 μg), streptomycin (10 μg), oxytetracycline (30 μg), gentamycin (10 μg), oxacillin (10 μg), erythromycin (15 μg), novobiocin (30 μg), amoxicillin (30 μg) were used in this current study. Each sample was examined with all antibiotics and 5 antibiotic discs were used for each A. hydrophila sample. All petri dishes were incubated at 37°C for 24 h after the placement of paper discs of antibiotics. A centimetre scale was used to measure the minimum inhibition of concentration (MIC) for all isolates.
Data analysis
Sequence data was analysed using Basic Local Alignment Search Tool (BLAST) of National Center for Biotechnology Information (NCBI), USA. The antibiotic sensitivity data was analysed using MS Excel and presented in tabular and graphical forms.
Acknowledgements
This research was undertaken through the project “Occurrence of epizootic ulcerative syndrome (EUS) and its impact on the biodiversity status of small indigenous species (SIS) in beels of Sylhet region ” -
funded by Sylhet Agricultural University Research System(SAURES)
under Fund for University Teachers Under UGC Research Grants for 2020-2021.
Novelty Statement
This study is a noble research for bacteria identification using both traditional and molecular methods, including its antibiotic sensitivity.
Author’s Contribution
All authors contributed equally.
Conflict of interest
The authors have declared no conflict of interest.
REFERENCES

Supplementary Figure 1: Aeromonas hydrophila colonies grown on Rimler- Shotts (RS) selective medium.
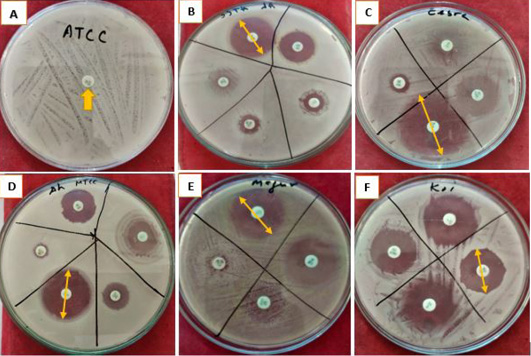
Supplementary Figure 2: Inhibition zone resulted using different antibiotics in pure culture of five A. hydrophila strains/isolates. Letters: A, Antibiotic sensitivity of A. hydrophila Strain ATCC 36562; B, Antibiotic Sensitivity of ATCC 36562; C, Antibiotic sensitivity of A. hydrophila isolated from Zebra fish (Danio rerio); D, Antibiotic sensitivity of Strain MTCC 1739; E, Antibiotic sensitivity of A. hydrophila isolated from walking catfish (Clarias batrachus); F, Antibiotic sensitivity of A. hydrophila isolated from Koi Carp (Cyprinus rubrofuscus). Yellow arrows showing the inhibition zone measured in (cm).





