Advances in Animal and Veterinary Sciences
Research Article
Advances in Animal and Veterinary Sciences 2 (2): 116 – 119Comparison of D–Loop Gene for Subspecies Confirmation of Tigers (Panthera Tigris)
Alisha Alisha, Prasad Minakshi*, Koushlesh Ranjan, Gaya Prasad,
*Corresponding author: minakshi.abt@gmail.com
ARTICLE CITATION:
Alisha A, Minakshi P, Ranjan K, Prasad G (2014). Comparison of D–loop gene for subspecies confirmation of tigers (panthera tigris). Adv. Anim. Vet. Sci. 2 (2): 116 – 119.
Received: 2013–12–12, Revised: 2014–01–10, Accepted: 2014–01–11
The electronic version of this article is the complete one and can be found online at
(
http://dx.doi.org/10.14737/journal.aavs/2014/2.2.116.119
)
which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
ABSTRACT
Tiger (Panthera tigris), the biggest cat species and the top predator has fallen into endangered category. In the present study, we confirmed the subspecies of tiger with help of DNA based methods. The mitochondrial control region gene (D–loop) sequence was utilized for this purpose. DNA was extracted from tiger blood sample originally obtained from mini zoo of Rohtak, Haryana. The D–loop sequence was amplified and submitted to GenBank with accession number HM362438. Based on D–loop sequence identity and phylogenetic analysis Panthera tigris tigris (Indian tiger) was differentiated from other Panthera tigris subspecies (Panthera tigris amoyensis, Panthera tigris corbetti, Panthera tigris sumatrae and Panthera tigris altacia) from different parts of the world. The present report also describes an easier, quicker and non hazardous protocol for the isolation of genomic DNA from tiger blood.
INTRODUCTION
The global adult tiger population as per Global Tiger Recovery Program (2012) has been estimated to be only 4,000 as compared to 1,00,000 in the beginning of this century (Chundawat et. al., 2012). The demographic distribution of tigers shows a great shrinkage in their range declining to only 7 percent of historic range (Dinerstein et. al., 2007). Forensic identification of tiger species can be carried out using various methods. However, molecular methods provide a direct and confirmatory approach to identify and distinguish among closely related species or subspecies (Gupta et. al., 2005). Also, the need to stop the ongoing havoc of poaching and illegal trade of tiger body parts rises the importance of DNA based molecular approach. It involves the use of both nuclear as well as mitochondrial (mt) DNA based markers. However, mt DNA markers have been reported to be more efficient than nuclear markers for identification and authentication of species (Rastogi et al., 2007), as the rate of evolution of mt genome is much faster than nuclear genome thereby helping in differentiation of closely related species (Brown et al., 1979; Kitpipit et. al., 2012). Phylogenetic relations can be easily drawn on the basis of the mt gene studies and thus, can be compared with the distant and near tiger relatives. The mt DNA control region (D–loop) analyses showed a central conserved region, two hypervariable segments along with size and sequence heteroplasmy in five species of the genus Panthera (Jae–Heup, et al., 2001). The control region of mt DNA (D–loop) was used successfully for species identification of five different species viz., red deer (Cervus elaphus), roe deer (Capreolus capreolus), fallow deer (Dama dama), mouflon (Ovis aries musimon) and wild boar (Sus scrofa) using hair samples (Parkanyi et al., 2013). The mt DNA control region (D–loop) was also used for phylogeographic patterns and evolution studies of two Neotropical cats viz. ocelot (Leopardus pardalis) and margay (Leopardus wiedii) (Eizirik et al., 1998).
Blood has been extensively used for obtaining genomic DNA (gDNA) for population studies; however, quicker, safer and easier protocols are needed, to apply them for larger population structure. Therefore, the present study was also planned to develop an easier and non – hazardous protocol for isolation of DNA from blood after doing certain modifications in the chelex gDNA extraction protocol (Walsh et. al., 1991).
MATERIALS AND METHODS
Sample Origin
The blood sample from tiger (named Saurabh) was received from Mini zoo of Rohtak District of Haryana state. The blood sample was sent in EDTA anticoagulant in the teaching veterinary clinics of the college for routine haemoprotozoan investigation.
Genomic DNA Extraction
The extraction of gDNA was done using standard protocol with certain modifications (Walsh et. al., 1991). Briefly, RBC lysis buffer containing 17mM Tris HCl (pH 7.65) and 140 mM NH4Cl, was added to the sample in the ratio of 1:2 and kept for 15 min at room temp. Centrifugation was done at 8,000 rpm for 20 min. RBC lysis step was repeated until a white pellet was obtained. The white pellet was dissolved in 100µl Lefton’s buffer (25mM Tris HCl, pH 7.5, 100mM EDTA and 1% SDS), mixed and kept for 15 min at room temp. To the lysate 200µl of 5% chelating resin, 2.5µl of proteinase K (10mg/ml) and 4.5 µl of DTT (2M) were added. This solution was incubated at 56°C for 45 minute with intermittent shaking. After incubation, the enzyme was inactivated in boiling water bath for 10 minute. Supernatant was collected in a fresh eppendorf tube after centrifugation at 12,000 rpm for 1 minute.
Quantification of gDNA
The extracted gDNA for further processing required more reliable values measured with a high fidelity spectrophotometer. The quality and quantity of gDNA was assessed using Picodrop™ high fidelity UV spectrophotometer (Astranet systems Ltd. ®, U.K.) as per standard procedure. In Brief, 1 µl of purified DNA sample was taken in the disposable UVpette® tips (Astranet systems Ltd ®, U.K.). The optical density of gDNA sample was determined at A260 and A280, keeping NFW as blank. The quantity of the gDNA was measured by using the standard reading of 1 O.D. at A260 for 50µg/ml. The reading was obtained in ng/µl.
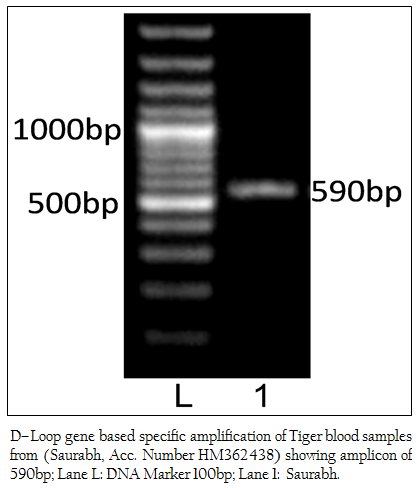
Figure 1: D–Loop gene based specific amplification of Tiger blood samples from (Saurabh, Acc. Number HM362438) showing amplicon of 590bp; Lane L: DNA Marker 100bp; Lane 1: Saurabh.
RRESULT
The genomic DNA from blood sample was extracted using chelex based method. The gDNA was found to be 143.16 ng/µl after measuring by Picodrop™ UV spectrophotometer. The gDNA was then subjected to D–loop gene specific PCR with standardized primer (Zhang et. al., 2006). The species specific D–loop gene primer pair yielded a PCR product of 590bp (Figure 1). The PCR amplicon of D–loop region was sequenced in our laboratory. The sequence data was submitted to GenBank data base and an accession number HM362438 was assigned to it. The sequence data of Saurabh (Accession number HM362438) showed more than 99% identity with Panthera tigris tigris after BLASTN+2.2.28 search (Zhang et al., 2000) in GenBank database. Pairwise nucleotide identity of Saurabh (Accession number HM362438) with other Panthera tigris subspecies were also calculate using Bioedit v 7.2.3 software (Hall, 1999) (Table 1). A D– loop gene based neighbor joining tree (NJ Tree) of Saurabh (Accession number HM362438) with other subspecies of tigers from different parts of the world was constructed using Mega 5.2 software (Tamura et al., 2011) (Figure 2).
DISCUSSION
Tiger subspecies confirmation can help in genome conservation in a better way and characteristics of different tiger subspecies can be maintained in the gene pool. The method that has been developed is easy, quick and most importantly requires few micro–liter of blood to yield a significant amount of DNA even from clotted and degraded blood sample, thus allowing further genetic analysis. As the mt DNA accumulates mutation faster than the nuclear DNA, it is helpful in studying the genetic diversity among the Indian tigers. The sequence analysis of D–loop from Saurabh tiger revealed that it was very closely related (>99% nucleotide identity) to Panthera tigris tigris from China and United Kingdom. However, it was also 98.9 to 97.6% identical with other Panthera tigris subspecies (Panthera tigris amoyensis, Panthera tigris corbetti and Panthera tigris sumatrae) from China, United Kingdom and USA. It showed the close relationship of Panthera tigris tigris subspecies and the common line of its evolution. However, it is distinct from other Panthera tigris subspecies from different parts of the world. Moreover, it was distantly related (only 40.7 to. number HM362438).
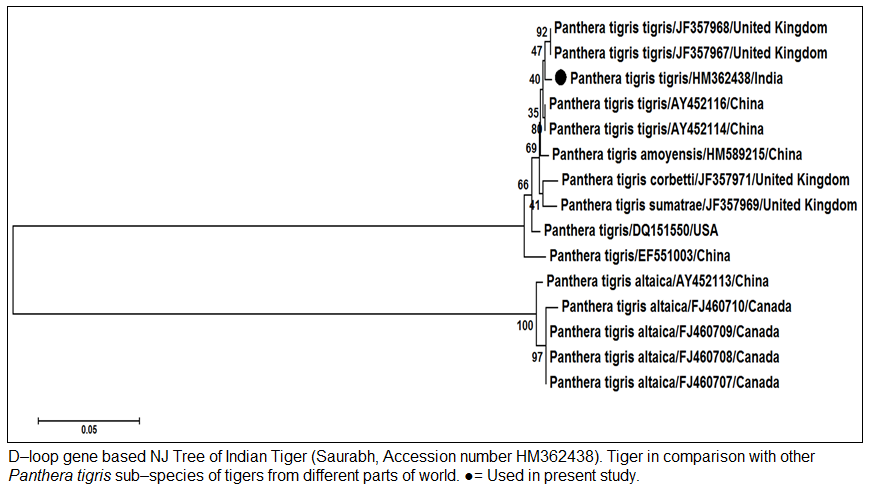
Figure 2: D–loop gene based NJ Tree of Indian Tiger (Saurabh, Accession Tiger in comparison with other Panthera tigris sub–species of tigers from different parts of world. Used in present study.
Panthera tigris altaica from China and Canada. The phylogenetic analysis also showed that Indian Tiger Saurabh (Accession number HM362438) formed a very close cluster with Panthera tigris tigris from China and United Kingdom. However, it was also closely related with different Panthera tigris subspecies (Panthera tigris amoyensis, Panthera tigris corbetti and Panthera tigris sumatrae) from China, USA and United Kingdom. The Panthera tigris altaica from China and Canada formed a separate cluster and were distantly related to Panthera tigris tigris. The D–loop gene along with other mitochondrial gene cytb can be used for the species differentiation of tiger and other wild animals. The species differentiation between Indian wolves and other Canid species were done using D–loop, 16S rRNA and cytb gene sequence (Aggarwal et. al. 2007).
CONCLUSION
Tiger is an endangered species in India and throughout the world. To control poaching, illegal trade of tiger body parts and pure line breeding in captive condition the proper species identification up to subspecies level is essential. In present study the control region of mitochondrial (D–loop) region was amplified, sequenced and compared with other Tiger subspecies for sub species confirmation. Our analyses showed that based on D–loop sequence Panthera tigris tigris (Indian tiger) can be differentiate from other Panthera subspecies.
ACKNOWLEDGEMENT
The research was funded by Department of Biotechnology, New Delhi. The authors are thankful to Department of Animal Biotechnology, LLR University of Veterinary and Animal Sciences, Hisar for providing infrastructural facility. We are also thankful to director of clinics Dr. Suresh Chander for providing sample.
CONFLICT OF INTEREST
We declare no conflict of interest.
REFERENCES
Aggarwal RK, Kivisild T, Ramadevi J, Singh L (2007). Mitochodrial DNA coding region sequences support the phylogenetic distinction of two Indian wolf species. J. Zool. Syst. Evol. Res. 45(2): 163 – 172.
http://dx.doi.org/10.1111/j.1439-0469.2006.00400.x
Brown WM, George M, Wilson AC (1979). Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA. 76: 1967 – 1971.
http://dx.doi.org/10.1073/pnas.76.4.1967
PMid:109836 PMCid:PMC383514
Chundawat RS, Habib B, Karanth U, Kawanishi K, Khan JA, Lynam T, Miquelle D, Nyhus P, Sunarto S, Tilson R, Sonam W (2011). Panthera tigris. In: IUCN 2012. IUCN Red list of threatened species. www.iucnredlist.org.
Dinerstein E, Loucks C, Wikramanayake E, Ginsberg J, Sanderson E, Seidensticker J, Forrest J, Bryja G, Heydlauff A, Klenzendorf S, Leimgruber P, Mills J, O'brien IG, Shrestha M, Simons R, Songer M (2007). The Fate of Wild Tigers. Biosciences. 57(6): 508 – 514.
http://dx.doi.org/10.1641/B570608
Eizirik E, Bonatto SL, Johnson WE, Crawshaw PG, Vie JC, Brousset DM, O'Brien SJ, Salzano FM (1998). Phylogeographic Patterns and Evolution of the Mitochondrial DNA Control Region in Two Neotropical Cats (Mammalia, Felidae). J. Mol. Evol. 47: 613 – 624.
http://dx.doi.org/10.1007/PL00006418
PMid:9797412
Global Tiger Recovery Program – Implementation report – June 2012. http://globaltigerinitiative.org/.
Gupta SK, Verma SK, Singh L (2005). Molecular insight into a wildlife crime: the case of a peafowl slaughter. Forensic Sci. Int. 154: 214 – 217.
http://dx.doi.org/10.1016/j.forsciint.2004.12.010
PMid:16182969
Hall TA (1999). BioEdit: a user–friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41: 95 – 98.
Jae–Heup K, Eizirik E, O'Brien SJ, Johnson WE (2001). Structure and patterns of sequence variation in the mitochondrial DNA control region of the great cats. Mitochondrion 14: 279 – 292.
http://dx.doi.org/10.1016/S1567-7249(01)00027-7
Kitpipit T, Tobe SS, A Linacre (2012). The complete mitochondrial genome analysisof the tiger (Pantheratigris). Mol. Bio. Rep. 39(5): 5745 – 5754.
http://dx.doi.org/10.1007/s11033-011-1384-z
PMid:22207170
Lopez JV, Cevario S, O'Brien SJ (1996). Complete nucleotide sequences of the domestic cat Felis catus) mitochondrial genome and a transposed mtDNA tandem repeat (Numt) in the nuclear genome. Genomics 33: 229 – 246.
http://dx.doi.org/10.1006/geno.1996.0188
PMid:8660972
Parkanyi V, Ondruska L, Vasicek D, Slamecka J (2013). Multilevel D–loop PCR identification of hunting game. Applied Translational Genomics. Article in Press.
Rastogi G, Dharne MS, Walujkar S, Kumar A, Patole MS, Shouche YS (2007). Species identification and authentication of tissues of animal origin using mitochondrial and nuclear markers. Meat Sci. 76 (4): 666 – 674.
http://dx.doi.org/10.1016/j.meatsci.2007.02.006
PMid:22061243
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evo. 28: 2731 – 2739.
http://dx.doi.org/10.1093/molbev/msr121
PMid:21546353 PMCid:PMC3203626
Walsh PS, Metzger DA, Higuchi R (1991). Chelex–100 as a medium for simple extraction of DNA for PCR based typing from forensic material. Biotechniques. 10: 506 – 513.
PMid:1867860
Zhang W, Zhang Z, Shen F, Hou R, Lv X, Yue B (2006). Highly conserved D–loop–like nuclear mitochondrial sequences (Numts) in tiger (Pantheratigris). J. Genet. 85(2): 107 – 116.
http://dx.doi.org/10.1007/BF02729016
PMid:17072079
Zhang Z, Schwartz S, Wagner L, Miller W (2000). A greedy algorithm for aligning DNA sequences. J. Comput Biol. 7(1–2): 203 – 214
http://dx.doi.org/10.1089/10665270050081478
PMid:10890397