Advances in Animal and Veterinary Sciences
Research Article
Antibiogram and Antibiotic Resistance Genes among Coagulase-Negative Staphylococci recovered from Bovine Mastitis
Mousa S. Walid1*, Akram A. Salama1, Shimaa S. Elnahriry2, Eman E. Abdeen2, Ghada Abd Elmonsef Hadad3, Usama H. Abo-Shama4
1Department of Animal Medicine and Infectious Diseases, Faculty of Veterinary Medicine, University of Sadat City, 32511, Egypt; 2Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, University of Sadat City, 32511, Egypt; 3Department of Animal Hygiene and Zoonoses, Faculty of Veterinary Medicine, University of Sadat City,32511, Egypt; 4Department of Microbiology and Immunology, Faculty of Veterinary Medicine, Sohag University, Sohag 82524, Egypt.
Abstract | Coagulase-negative Staphylococci (CoNS) have emerged as an important Staphylococci species implicated in bovine mastitis in dairy herds. This study was conducted to determine the prevalence, antibiogram, and antibiotic resistance genes among coagulase-negative Staphylococci recovered from cattle suffer from subclinical mastitis in Egypt. A total of 110 (36.7%) milk samples collected from 300 lactating cows were positive for the California mastitis test. On the Mannitol Salt Agar medium, out of 110 subclinical mastitis samples 62 (56.36%) were identified as CoNS isolates. Antibiotic sensitivity test conducted against nine types of antibiotics for CoNS strains that exhibited high susceptibility to most of the tested antibiotics, with particularly resistance pattern to oxicillin (41.9 %), and 11 (17.7%) CoNS isolate exhibited multidrug resistance (MDR). A total of 15 randomly selected isolates were subjected for detection of antibiotic-resistant genes among CoNS. The results indicated that mecA (73.3%) was the most identified gene, followed by tetK (60%) and ermB (13.3%) genes, whereas no detection for blaZ and vanA genes. In conclusion, our results indicate the importance of the regular surveillance of phenotypic and genotypic profiles of CoNS, isolates to ensure effective control measures and minimize the evolution of MDR strains and recommended the applying of antibiotic sensitivity test before treatment or random selection of antibiotics in field cases to avoid the emerging of resistance phenomena.
Keywords | Subclinical mastitis, CoNS, Antibiotic resistance.
Received | April 08, 2021; Accepted | April 30, 2021; Published | July 15, 2021
*Correspondence | Mousa S Walid, Department of Animal Medicine and Infectious Diseases, Faculty of Veterinary Medicine, University of Sadat City, 32511, Egypt; Email: walidsaadvet@yahoo.com
Citation | Walid MS, Salama AA, Elnahiriry SS, Abdeen EE, Hadad GAE, Abo-Shama UH (2021). Antibiogram and antibiotic resistance genes among coagulase-negative staphylococci recovered from bovine mastitis. Adv. Anim. Vet. Sci. 9(8): 1267-1274.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.8.1267.1274
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Walid et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Mastitis is defined as inflammations of the udder tissues resulting in sever reduction of milk yield and quality and causing significant economic losses in the cattle worldwide (Gezgen and Seker, 2016). Also, mastitis defined as an inflammatory condition associated with high somatic cell count (SCC) levels and drop in milk production (Taponen et al., 2007).
Staphylococci have been reported to be the most common bacterial cause of subclinical mastitis (Pitkälä et al., 2004). However, CoNS are typical common bovine pathogen involved in subclinical mastitis, particularly in heifer, and are generally associated with less severe signs (Taponen et al., 2006; Becker et al., 2014; Srednik et al., 2015). In most countries, Coagulase negative Staphylococci (CoNS) are progressively recognized as etiological agents associated with intramammary infections (Unal et al., 2012). Some studies presumed CoNS as emerged causative mastitis pathogens with major virulence determinants (Zhang and Maddox, 2000), with a high degree of resistance to antimicrobials (Rajala-Schultz et al., 2009), and the capability to cause chronic infectious diseases (Gillespie et al., 2009). Several strains of CoNS species have been implicated in intramammary infections, although the most frequently isolated CoNS from bovine mastitis are S. chromogenes, S. simulans, S. xylosus, S. epidermidis, S. hyicus, and S. haemolyticus (Thorberg et al., 2009; Park et al., 2011). Due to the increasing importance of CoNS in bovine mastitis, the identification of CoNS species is necessary to design effective control approaches for CoNS mastitis (Sawant et al., 2009).
Treatment with antimicrobial agents is the most widely used protocol for the treatment and control of mastitis (Gomes and Henriques, 2016). However, the excessive and abuse of antibiotics in humans and animals’ practices is strongly linked to the evolution of antibiotic resistance against several antimicrobial groups particularly in veterinary medicine (Schwarz et al., 2018). Also, CoNS may have a role in the transmission of resistance genes (Becker et al., 2014), and emerging of new multidrug resistant strains (Otto, 2012; Vitali et al., 2014). In recent study Qu et al. (2019) reported that CoNS harbored various antimicrobial resistance genes such as mecA, tetK, tetL, tetM, dfrG, and ermB which are responsible for methicillin, tetracycline, trimethoprim and erythromycin respectively.The constant surveillance and monitoring of the antimicrobial resistance profiling of the CoNS isolates will provide valuable database about the efficacy of the antibiotics in control of mastitis and manipulative an effective treatment and control measures for bovine mastitis (Veras et al., 2008). As result of the limited available information about the phenotypic and genotypic resistance profile of CoNS strains isolated from subclinical mastitis in Egypt as recent reported by (Nayel at el., 2020) who revealed the emerging of CoNS in clinical mastitis and subclinical mastitis with prevalence 60% and 67.27% respectively in Egyptian cows with high resistance against penicillin and oxacillin in Menofiuya Governorate. Therefore, this study was planned to investigate the phenotypic and molecular detection of antibiotics resistance genes in CoNS strains from bovine mastitis, Egypt.
Material and methods
Sample collection and study area
A total of 300 mixed breed dairy cows aged from 3-7 years old from individual cases from Sadat City, Menoufiya Governorate, Egypt, were examined during the period from December 2019 to March 2020. One hundred and ten cows showed positive reaction with California mastitis test (CMT), which indicated subclinical mastitis. CMT was performed according to the methodology described by (Schalm and Noorlander, 1957). The CMT-positive milk samples were collected aseptically and transferred under cold conditions (4°C) at the earliest possible to the laboratory for bacterial isolation and identification.
Bacteriological isolation and identification
For the isolation of Staphylococci, the milk samples were centrifuged and the sediment was then cultured in Mannitol Salt Agar (Oxoid Ltd. UK) and incubated for 1–2 days at 37°C. Confirmatory identification of CoNS was implemented by Gram staining then subjected to catalase and finally by tube coagulase test (Tortora et al., 2013). Pink color colonies were considered as CoNS which were confirmed by standard biochemical activities according to (Murray et al., 2003). Evaluation of hemolytic activity on sheep blood agar (5-7%) was performed as described by (Quiblier et al., 2011). Biofilm activity on Congo red agar medium (Arciola et al., 2015). The positive Congo red activities produced black colonies while non-producer gave red colonies.
Antibiogram profile of CoNS isolates recovered from bovine mastitis
The identified CoNS isolates were subjected for in vitro using the disk diffusion method as described by (CLSI, 2017) against different antibiotic groups to determine the susceptibility and resistance pattern to nine antimicrobial disks (Oxoid Ltd.). The used antibiotics: penicillin: P (100 IU), oxacillin: OX (1 μg), amoxicillin/clavulanic acid: AMC (30 μg), vancomycin: VA (30 μg), erythromycin: E (15 μg), gentamycin: CN (10 μg), chloramphenicol: C (30 μg), tetracycline: TE (30 μg) and ciprofloxacin: CIP (5 μg). The results were recorded as resistant, intermediate, or susceptible according to the diameter of the inhibitory zone as established by the Clinical and Laboratory Standards Institute (CLSI, 2017).
Molecular characterization of antibiotic resistance genes in CoNS isolates
Freshly grown typical CoNS colonies were harvested, and DNA extraction was performed according to the manufacturer’s guidelines using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany).
Table 1: The PCR primers for CoNS antibiotic resistance genes.
| Target gene |
Primer sequences
|
Amplified amplicon (bp) | Primary denaturation | Secondary denaturation | Annealing |
Extension
|
Final extension |
|
mecA |
GTAGAAATGACTGAACGTCCGATAA CCAATTCCACATTGTTTCGGTCTAA | 310 |
94˚C 5 min. |
94˚C 45 sec. |
50˚C 45 sec. |
72˚C 45 sec. |
72˚C 10 min. |
|
blaZ |
ACTTCAACACCTGCTGCTTTC TGACCACTTTTATCAGCAACC | 173 |
94˚C 5 min. |
94˚C 30 sec. |
54˚C 30 sec. |
72˚C 30 sec. |
72˚C 7 min. |
|
tetK |
GTAGCGACAATAGGTAATAGT GTAGTGACAATAAACCTCCTA | 360 |
94˚C 5 min. |
94˚C 30 sec. |
54˚C 40 sec. |
72˚C 40 sec. |
72˚C 10 min. |
|
ermB |
CATTTAACGACGAAACTGGC GGAACATCTGTGGTATGGCG | 425 |
94˚C 5 min. |
94˚C 30 sec. |
51˚C 40 sec. |
72˚C 45 sec. |
72˚C 10 min. |
|
vanA |
CATGACGTATCGGTAAAATC ACCGGGCAGRGTATTGAC |
885 |
94˚C 5 min. |
94˚C 30 sec. |
56˚C 41 sec. |
72˚C 50 sec. |
72˚C 10 min. |
All genes were amplified for 35 cycles
Molecular detection of CoNS antimicrobial resistance genes was performed in a 25-µl reaction mixture containing 12.5 µl of Emerald Amp Max PCR Master Mix (Takara, Japan), 1 µl of each primer at 20 pmol concentration, 4.5 µl of water, and 6 µl of DNA template. Table 1 lists the primers (Metabion,, Germany) used for detecting the genes encoding the methicillin resistance (mecA) (McClure et al., 2006), β-lactam resistance (blaZ) (Duran et al., 2012), tetracycline resistance (tetK) (Duran et al., 2012), vancomycin resistance (vanA) (Patel et al., 1997), and macrolide resistance (ermB) (Schlegelova et al., 2003).
The PCR reaction was applied in an Applied Biosystem 2720 thermal cycler (Applied Biosystems, Foster, CA). A total 15 µl aliquots of all PCR products and Gelpilot 100 bp (Qiagen, Germany) were loaded 1.5% agarose gel electrophoresis (Applichem, Germany). The gel was photographed by a gel documentation system (Alpha Innotech, Biometra), and the data were analyzed through computer software (The CFR 21 Part 11).
RESULTS
Prevalence of CoNS recovered from subclinical mastitis cases
Of the total 300 dairy cows examined in this study, 110 (36.7%) were diagnosed with subclinical mastitis depending on the CMT result. Among the CMT-positive samples, 62 were found to be positive on the Mannitol Salt Agar medium (56.36%) CoNS isolates as showed in Table (2).
Based on the biochemical and enzymatic activity, CoNS strains were serotyped into S. epidermidis, S. saprophyticus, S. heamolyticus with prevalence 30 (48.4%), 20 (32.3%), 12 (19.4%) respectively. All types of CoNS species were positive for catalase activity, while 12 (19.6%) isolates from
Table 2: Prevalence of CoNS recovered from subclinical mastitis cases
| Total examined animals | Positive samples for CMT | Positive CoNS | ||
|
300 |
No | % | No | % |
| 110 | 36.7 | 62 | 56.36 | |
S. heamolyticus exhibited only α-hemolysis, whereas none of the CoNs species exhibited DNase activity. In addition, only 5 (8.19%) CoNS species were positive for biofilm activities. Concerning to trehalose fermentation the S. epidermidis was the only CoNS species produce a positive test. The sensitivity to novobiocin revealed that S. epidermidis and S. heamolyticus showed sensitivity to novobiocin while S. saprophyticus showed resistance for novobiocin.
Phenotypic resistance of CoNS isolates recovered from bovine mastitis
Sensitivity test against nine antibiotics for 62 CoNS species revealed high susceptibility to gentamycin, ciprofloxacin, amoxicillin/clavulanic acid, chloramphenicol, tetracycline, penicillin, vancomycin, and erythromycin, with susceptibility rates of 98.39%, 96.77%, 91.94%, 90.32%, 75.8%, 74.2%, 72.58%, and 72.58%, respectively. Meanwhile, the resistance pattern of CoNS species 41.9% for oxacillin (Table 3).
Multidrug resistance (MDR) profiles of CoNS species recovered from subclinical mastitis.
Among the 62 CoNS species, 11 (17.7%) exhibited MDR to three to four groups of antibiotics. Of these, only 1 isolate (1.6%) exhibited MDR to four antibiotic groups, in
Table 3: Antibiogram pattern of CoNS recovered from subclinical mastitis cases
| Antimicrobial classes | Antimicrobial agents | No of CoNS isolates (%) | |||||
| R | % | I | % | S | % | ||
| Beta –lactams | Oxacillin (OX) | 26 | 41.94 | 4 | 6.45 | 32 | 51.61 |
| Penicillin (P) | 12 | 19.35 | 4 | 6.45 | 46 | 74.2 | |
| Amoxicillin/Clavulanic acid (AMC) | 5 | 8.06 | 0 | 0 | 57 | 91.94 | |
| Tetracycline | Tetracycline (TE) | 13 | 20.97 | 2 | 3.23 | 47 | 75.8 |
| Macrolides | Erythromycin (E) | 15 | 24.19 | 2 | 3.23 | 45 | 72.58 |
| Fluoroquinolones | Ciprofloxacin (CIP) | 2 | 3.23 | 0 | 0 | 60 | 96.77 |
| Chloramphenicol | Chloramphenicol (C) | 6 | 9.68 | 0 | 0 | 56 | 90.32 |
| Aminoglycosides |
Gentamycin (CN) |
1 | 1.61 | 0 | 0 | 61 | 98.39 |
| Glycopeptides | Vancomycin (VA) | 2 | 3.23 | 15 | 24.19 | 45 |
72.58 |
R=Resistance I= Intermediate S=Sensitive
Table 4: Multidrug resistance profiles of CoNS recovered from subclinical mastitis
| CoNS resistant isolates |
Resistance profile |
No. of resistance antimicrobial classes |
||||
| No. | %* | |||||
| 1 | 1.61 |
|
E-CIP-TE-C | 4 | ||
| 1 | 1.61 |
|
OX-P-E-C | 3 | ||
|
1 |
1.61 |
|
OX-AMC-E-CN | 3 | ||
| 1 | 1.61 |
|
OX-TE-C | 3 | ||
| 2 | 3.23 |
|
OX-TE-E | 3 | ||
| 2 | 3.23 |
|
TE-C-E | 3 | ||
| 2 | 3.23 |
|
OX-P-E-C | 3 | ||
| 1 | 1.61 |
|
OX-TE-VA | 3 | ||
OX=Oxacillin P=Penicillin AMC=Amoxicillin/Clavulanic acid E=Erythromycin
CIP=Ciprofloxacin TE=Tetracycline C=Chloramphenicol CN=Gentamycin VA=Vancomycin
*Percentage was estimated according to the total number of isolates 62 for CoNS.
addition to 10 isolates (16.1%) showing MDR to three antibiotic groups (Table 4).
Molecular detection of CoNS antimicrobial resistance
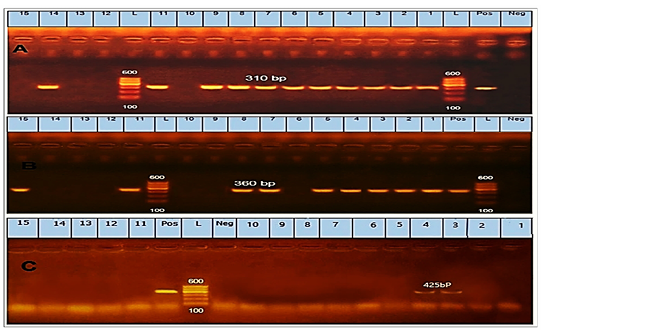
Regarding to the prevalence of antibiotic-resistant, mecA was the most detected gene in 11 (73.3%) isolates of CoNS (Fig.1A). Furthermore, the blaZ not detected at any of the tested CoNS isolates. Regarding the tetK gene, it was detected in 9 isolates with a prevalence of 60% in CoNS (Fig.1B). The ermB gene was identified in 2 isolates with a prevalence of 13.3% in CoNS (Fig. 1C).
Phenotypic resistance and antibiotic resistance genes among MRCoNS species
Table 4 shows the distribution and the correlation between the presence of phenotypic patterns, and antibiotic resistance genes among (MRCoNS) species isolated from subclinical mastitis. Furthermore, 8/15 (53.3%) among methicillin-resistant CoNS (MRCoNS) exhibited MDR in the phenotypic test and carried (two to three) antibiotic resistance genes.
Table 5: The phenotypic resistance and antibiotic resistance among MRCoNS strains recovered from subclinical mastitis
| Strains No. | Phenotypic resistance | Biochemical activities | Antibiotic resistance genes | ||
| Coagulase | Haemolysis | Biofilm activity | |||
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 | OX-P-E-TE-C-CIP OX-P-E-CIP-TE-VAN OX-P-AMC-E-TE-C OX-TE-C-E-AMC OX-P- TE-E-CN-C-AMC OX- E- OX-P- TE-E- VAN-AMC OX-P-TE-E OX-P- E OX-P- E OX-P-E-TE-C- AMC OX-P-E OX-P-E OX-P-E OX-TE-E | - - - - - - - - - - - - - - - | Α α α α α α α α α α α α α α | + + - - + - + - - - + - - - - | tetK, mecA tetK, mecA tetK, mecA, ermB tetK, mecA, ermB tetK, mecA mecA tetK, mecA, tetK, mecA, mecA ND tetK, mecA ND ND mecA tetK |
Bovine mastitis is the most economic and productive disease affecting the livestock industry which can be graded as clinical or sub-clinical mastitis (Krishnamoorthy et al., 2017). A large drop in milk production due to subclinical mastitis is a major financial loss (Romero et al., 2018).
In this study, the overall prevalence of subclinical mastitis was 36.7%. Nearly similar findings were reported by (Mekebib et al., 2010; Workineh et al., 2002; Sukur and Esendal, 2020; Ayano et al., 2013) 34.8 % 38.2 %, 39.1% and 41.02% respectively. On other hand, higher prevalence rate 62.6% and 50.4% were reported by (Mpatswenumugabo et al., 2017; Abebe et al., 2016). Lower prevalence rate 26.7% and 28.5% was recorded by (Abdel-Tawab et al., 2016; Kayesh et al., 2014) in Egypt, and Bangladesh, respectively. These differences in subclinical mastitis prevalence rates may be attributed to the differences in the animal’s breeds, management strategies, and the applicable health practices (Rathod et al., 2017).
The present study revealed that CoNS species was isolated in 62 (56.63%) among the CMT-positive samples and S. epidermidis, S. saprophyticus, S. heamolyticus were the detected CoNS species with 30 (48.4%), 20 (32.3%), 12(19.4%) respectively. Our results agree with those of several scientific reports that described CoNS as a major bacterial etiology connected in bovine mastitis. For example, (Dieser et al., 2013; Zigo et al., 2017) reported that CoNS was the most isolated bacterial group from milk samples of dairy cows with subclinical mastitis. Furthermore, (Zigo et al., 2019) reported that CoNS accounted for 43.4% and 50.0% in two cattle herd examined for mastitis. In contrast to our study, (Mahmoud et al., 2015) recorded a lower percentage (8.9%) of CoNS isolates from subclinical mastitis samples in Egypt. In a recent comparative study, (Sukur and Esendal, 2020) reported that S. chromogenes, S. capitis and S. simulans were the most prevalent CoNS species in subclinical mastitis with (41.2%), (14.7%) and (11.8%) respectively in North Cyprus. This was also supported by (Vanderhaeghen et al., 2015) who demonstrated that S. chromogenes, S. haemolyticus, S. epidermidis, S. simulans and S. xylosus are common isolated CoNS species isolated from bovine mastitis. Likewise, (Nayel et al., 2020) isolated S. epidermidis and S. saprophyticus from bovine clinical mastitis in Egypt.
In the current study, the antibiotic resistance profile of the 62 CoNS species that extremely showed susceptibility to gentamycin, ciprofloxacin, amoxicillin/clavulanic acid, chloramphenicol, tetracycline, penicillin, vancomycin, and erythromycin. Meanwhile, the resistance pattern of CoNS species against the oxacillin 41.94% was observe. This is not consistent with the result reported by (Phophi et al., 2019) who recorded that 90% of CoNS were resistant to at least one antimicrobial and 51% showed MDR, and the highest resistance against ampicillin (90%) and penicillin (89%). A low resistance (19.35%) against penicillin among CoNS species was observed in our study compared to that reported in mastitis of dairy cattle in Estonia (38.5%) (Pitkälä et al., 2007), Finland (32%) (Simojoki et al., 2012), and South Africa (63%) (Phophi et al., 2019).
Vancomycin resistance is very important due to its use in the treatment of MRSA cases, and in this study, the uncommon resistance of vancomycin among CoNS strains was (3.23%), which is in parallel to the study of (Phophi et al., 2019), who observed no vancomycin resistance among CoNS strains recovered from mastitis in dairy cattle as well as was report of (Bengtsson et al., 2009) in Sweden revealed the resistance of CoNS to β-lactams antibiotics. The low resistance of CoNS strains to vancomycin may be attributed to the uncommon use of antibiotics such as vancomycin in systemic bovine mastitis as injectable drug but only used as local intrammamary infusion.
With reference to CoNS, 11/62 (17.74%) exhibited MDR to three to four groups of antibiotics. Previously similar findings have been described in several countries, including the USA, Switzerland, and the Netherlands (Sawant et al., 2009; Moser et al., 2013; Sampimon et al., 2011). Moreover, (Huber et al., 2011) found that 33%–49% of all 414 CoNS strains exhibited MDR to several antimicrobial groups. Moreover, (Van Duijkeren et al., 2004) reported MDR in CoNS strains isolated from animals with clinical diseases.
Molecular surveillance of the five antibiotic-resistant genes (mecA, blaZ, ermB, tetK, and vanA), our results showed that mecA, tetK, and ermB were the common prevalent resistance genes among the tested with prevalence rates 73.33%, 60%, and 13.3%, respectively. Meanwhile, blaZ and vanA genes were not detected. This result contrasted with the study of (Qu et al., 2019) who detected high prevalence rates of blaZ (100%), mecA (73%), tetK (79%), and tetM (96%) among the CoNS isolates from bovine mastitis. Additionally, (Frey et al., 2013) successfully detected mecA, blaZ, tetK, and ermB with prevalence rates of 9.7%, 90.7%, 95.4%, and 7.4%, respectively. In Lithuania, (Klimiene et al., 2016) reported the prevalence rates of blaZ, tetK, and mecA genes as 66.6%, 38.1%, and 23.8%, respectively.
Concerning MRCoNS, 53.33% of the strains exhibited MDR in the phenotypic testing and possessed (two to three) antibiotic resistance genes. Correspondingly, (Frey et al., 2013) reported that CoNS isolated from bovine mastitis samples exhibited MDR profiles. Likewise, (Araujo et al., 2006) reported that biofilm-producing methicillin-resistant S. epidermidis isolates exhibited MDR compared with non-producer isolates.
Conclusion
In conclusion, this study concluded that CoNS were considered an emerging cause of subclinical intramammary infection in individual bovine mastitis cases. Moreover, the higher predominance of phenotypic and genotypic resistance of CoNS species in this study indicates the potential economic loss. Therefore, it can recommend that the hygiene regimen and regular investigation for MDRCoNS screening and recommended the significance for antibiotic sensitivity test to minimize the emerging of resistance CoNS strains in subclinical mastitis in cows in the studied area.
Acknowledgements
The authors would like to thank the staff members of Animal Medicine and Infectious Diseases, Faculty of Veterinary Medicine, University of Sadat City for the help for finishing the manuscript.
Conflict of interest
The authors have no conflict of interest.
authors contribution
WSM, UHA, E E A, AAS, S S E, and G E H involved in the conception of the research idea and methodology design, performed data analysis and interpretation, laboratory work, and prepared the manuscript, and these authors contributed equally to the manuscript, and contributed in the samples collection and laboratory work. All authors read and approved the final manuscript.
References