Advances in Animal and Veterinary Sciences
Research Article
Characterization of Toxin Gene Profiles and Antibiotic Resistance Genes of Methicillin Resistant Staphylococcus aureus Isolated from Ducks
Mona. A. A. AbdelRahman1, Fatma Amer2
1Department of Bacteriology, Reference Laboratory for Veterinary Quality Control on Poultry Production, Animal Health Research Institute, Agriculture Research Center, P.O. Box 264, Dokki, Giza 12618, Egypt; 2Department of Biotechnology, Reference Laboratory for Veterinary Quality Control on Poultry Production, Animal Health Research Institute, Agriculture Research Center, P.O. Box 264, Dokki, Giza 12618, Egypt.
Abstract | Although Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most widespread Bacteria causing infection over the world for poultry sector, MRSA in duck farms hasn’t yet been investigated seriously. Thus, this study intensively investigates the prevalence of MRSA through 100 duck farms. S. aureus has been isolated from 21 farms. The classical enterotoxins (A–E) production plus toxic shock syndrome toxin (TSST-1) were screened by PCR, only four strains harbored sed gene. The antimicrobial resistance profiles were tested by agar diffusion assays showed that 90.5% of isolates exhibit multidrug resistance (MDR). All the tested isolates revealed 100% resistance towards penicillin G., ampicillin and cefoxitin. However, the resistances to kanamycin, tetracycline and gentamycin were 90.5%, 85.7% and 81% respectively. Additionally, Methicillin-resistant S. aureus (MRSA) isolates were identified by the presence of mecA gene, but all isolates were negative for mecC gene. The isolates were tested for the presence of four antimicrobial resistance genes including tetracycline resistance gene tetK that was detected in all isolates. Secondly, 90.5% of isolates carried erythromycin resistant gene ermB as well as gentamicin resistant gene, aacA-aphD. Finally, none of those isolates has carried vanA gene for vancomycin resistance. Consequently, continuous tracking the presence of MRSA in duck farms is very important to avoid developing a reservoir for antimicrobial resistances.
Keywords | Staphylococcus aureus, tetK, Duck, mecA, Enterotoxins.
Received | February 06, 2021; Accepted | March 17, 2021; Published | July 01, 2021
*Correspondence | Mona AA AbdelRahman, Department of Bacteriology, Reference Laboratory for Veterinary Quality Control on Poultry Production, Animal Health Research Institute, Agriculture Research Center, P.O. Box 264, Dokki, Giza 12618, Egypt; Email: drmonaali2000@yahoo.com
Citation | AbdelRahman MAA, Amer F (2021). Characterization of toxin gene profiles and antibiotic resistance genes of methicillin resistant staphylococcus aureus isolated from ducks. Adv. Anim. Vet. Sci. 9(8): 1150-1158.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.8.1150.1158
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 AbdelRahman and Amer. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
S. aureus is one of the most common bacterial diseases in poultry which causes drop in egg production, body weight loss, and lameness which is leading to carcass condemnation. Therefore, S. aureus has negative impact on the economy based on farming business alongside the foodborne illness causes (Andreasen, 2013). Moreover, S. aureus influences on the human health and causes food poisoning regarding to their enterotoxins productions (Han et al., 2013).
Although diagnosing, detecting and genotyping of S. aureus are routinely processed among sick poultry (Andreasen , 2013) , alongside pigs, cows and goats, (Wang et al.,2017), the detection of S. aureus in ducks is too rare. However, in Egypt, the ducks are taking the second place of the poultry meat production (Radwan et al., 2019). On the other hand, the S. aureus is considered as one of the bacteria causing diseases in ducks and is responsible for suppurative dermatitis, suppurative arthritis, and septicemic lesions (Smyth and McNamee, 2008). This is in a similar way to chicken but it has different immunity response (Andreasen, 2013; Eid et al., 2019). Moreover, the S. aureus causes diseases in both human and animals as consequences of virulence factors production also (Marek et al.,2018). These virulence factor are such as the enterotoxins which are heat stable, having 23 types including main fives types namely SEA, SEB, SEC, SED, and SEE. Additionally toxic shock syndrome of toxin-1 (TSST-1) (Wang et al., 2017; Ono et al., 2015) is one of virulence factors as well.
Methicillin-resistant S. aureus (MRSA) has the ability to be transmitted to human from livestock and vice-versa (Marek et al., 2018), so that it causes a severe hazardous effect on the public health. MRSA carries variable antibiotic resistance patterns leading to the treatment failure and also causes nosocomial infection in some cases (Han et al., 2013; Ben Zakour et al., 2008; Achek et al., 2018). Evermore, Methicillin-resistant S. aureus (MRSA) is characterized by harboring mecA gene which is responsible for methicillin resistance. Therefore, the gene encrypts against penicillin-binding protein (PBP2a) which is less influenced by beta-lactam antibiotics. However this encoded gene is located at the staphyloccoccal cassette chromosome mec(SCCmec) part, which is a genomic island ubiquitously disseminated among staphylococci (Han et al., 2013; García-Garrote et al., 2014). Recently, in Germany, a new gene has been discovered and known as mecC gene but its similarity to mecA gene is about 70% of identical nucleotide (Stegger et al., 2012), noting that both animals and humans are carrying mecC gene (García-Garrote et al., 2014).
Thus, MRSA has been categorized as one of a superbug (Dweba et al., 2019; WHO, 2020), because of its wide spread alongside to the misusage of antibiotics treating S. aureus infections which both are developing the antibiotic resistance. Throughout livestock and food producing animals the S. aureus infection can be easily transmitted to human via the food chain (Wang et al., 2017; WHO, 2020; Rodríguez-Lázaro et al., 2017; Grema et al., 2015). The resultant, several cases had been reported as S. aureus infections in human (Rodríguez-Lázaro et al., 2017; Blumental et al., 2013).
Erythromycin is one of the antibiotics used in the treatment of Gram-positive bacteria as S. aureus (Khan et al., 2002). However, there are about 17 genes responsible for erythromycin resistance in S. aureus (Nawaz et al., 2000), where the most common genes are ermA, ermB and ermC. These genes are targeting and responsible for ribosomal manipulation in macrolides, lincosamides and type B streptogramins (Achek et al., 2018). Additionally, both tet(K) and tet(L) genes are rising up the resistance for tetracyclines at staphylococci (Wendlandt et al., 2013).
Therefore, the aim of this study was MRSA detection from apparently healthy duck farms with their phenotypic and genotypic antimicrobial resistance characters as well as enterotoxin genes.
Materials and methods
Samples
The Samples were taken from 100 duck farms, where 3 freshly dead ducks were transferred in ice boxes to reference laboratory for veterinary quality control on poultry production within 24 h for examination and testing. All samples were collected from ducks joint while the stab swabs were gathered from internal organs such as lung and liver.
Bacterial Isolation
The swabs from lung and liver organs were inoculated into 5% sheep blood agar and incubated for 24 h at 37°C. After incubation, the S. aureus isolates are ß-hemolytic. Heavily contaminated material was inoculated into a selective medium inhibitory for gram-negative bacteria, such as mannitol-salt agar (Andreasen, 2013). After that, the typical Staphylococcus spp. colonies were investigated and examined by gram staining, slide catalase test, oxidase test, and tube coagulase test (Quinn et al., 2002).
Antimicrobial Susceptibility Tests
Antimicrobial susceptibility testing for 21 S. aureus isolates throughout 18 antimicrobials agents was performed by using the disc diffusion method. The test was performed according to the Clinical and Laboratory Standards Institute (CLSI, 2015). The 18 antibiotics used in the disc diffusion method are namely: (Oxoid® ) Penicillin-G 10 I.U (P10); cefoxitin 30 μg (FOX30); Oxacillin 1µg (OX1); gentamicin 10 μg (CN10); Kanamycin 30 μg (K30); Ciprofloxacin 5 µg (CF5); clindamycin 2 μg (DA2); Azithromycin 15 μg (AZM15); erythromycin 15 μg (E15 ); Chloramphenicol 30 µg (C30); trimethoprim/sulfamethoxazole 1.25-23.75µg (SXT), doxycycline 30µg (DO30); Tetracycline 30 µg(T30), amoxicillin + clavulanic acid 20 + 10 μg (AMC30); cefotaxime 30µg (CTX 30); vancomycin 30 µg(VA 30); levofloxacin 5µg(Lev 5), ampicillin 10 μg (AMP10).
According to CLSI guidelines, after aerobic incubation at 37°C for 18-24 h, the susceptibilities of S. aureus isolates to the individual antimicrobial agents were determined and interpreted. The Test results were considered valid only, when the diameters of the inhibition zones for the control S. aureus (ATCC 25922) strain were within the performan-
Table 1: PCR primers used in this study and sizes of PCR products of methicillin resistance, enterotoxins (A-E), TSST-1, and antibiotic resistance genes.
| Target | Gene | Primer Sequence 5'-3' | Amplified fragment | References |
|
Methicillin resistance |
mecA | 5’GTAGAAATGACTGAACGTCCGATAA3’ | 310 bp | (Nowrouzian et al.,2013) |
|
5’CCAATTCCACATTGTTTCGGTCTAA3’ |
||||
|
mecC
|
5’GAAAAAAAGGCTTAGAACGCCTc3’ | 138 bp |
(Stegger et al.,2012) |
|
| 5’GAAGATCTTTTCCGTTTTCAGC3’ | ||||
| Enrerotoxins | sea | 5’GGTTATCAATGTGCGGGTGG3’ | 102 bp |
(Betley and Mekalanos, 1988) |
| 5’CGGCACTTTTTTCTCTTCGG3’ | ||||
| seb | 5’GTATGGTGGTGTAACTGAGC 3’ | 164 bp |
(Jones and Khan, 1986) |
|
| 5’CCAAATAGTGACGAGTTAGG 3’ | ||||
| sec | 5’AGATGAAGTAGTTGATGTGTATGG 3’ | 451 bp |
(Bohach and Schlievert, 1987) |
|
| 5’CACACTTTTAGAATCAACCG 3’ | ||||
| sed | 5’CCAATAATAGGAGAAAATAAAAG 3’ | 278 bp |
(Bayles and Iandolo , 1989) |
|
| 5’ATTGGTATTTTTTTTCGTTC 3’ | ||||
| see |
5’AGGTTTTTTCACAGGTCATCC 3’ |
209 bp | Couch et al.,1988 | |
| 5’CTTTTTTTTCTTCGGTCAATC 3’ | ||||
| Toxic shock syndrome | tsst-1 | 5’ACCCCTGTTCCCTTATCATC 3’ | 326 bp |
(Blomster-Hautamaa et al.,1986) |
| 5’TTTTCAGTATTTGTAACGCC 3’ | ||||
|
Erythromycin resistance |
erm(B)-1 erm(B)-2 |
5’CATTTAACGACGAAACTGGC 3’ | 425 bp |
(Jensen et al.,1999) |
|
5’GGAACATCTGTGGTATGGCG 3’ |
||||
| Tetracycline resistance | tetK | 5’GTAGCGACAATAGGTAATAGT 3’ | 360 bp |
(Strommenger et al.,2003) |
| 5’GTAGTGACAATAAACCTCCTA 3’ | ||||
|
Aminoglycoside resistance |
Aac6-aph2 | 5’GAAGTACGCAGAAGAGA 3’ | 491 bp |
(Choi et al.,2003) |
| 5’ACATGGCAAGCTCTAGGA 3’ | ||||
| Vancomycin resistance | vanA | 5’CATGACGTATCGGTAAAATC 3’ | 885 bp |
(Patel et al., 1997) |
-ce ranges.
Molecular Identification of Staph Aureus Isolates |Using Polymerase Chain Reaction Assay:
DNA extraction was fulfilled using QIAamp DNA mini kit (Qiagen, Germany, GmbH) according to manufacturer’s instruction.
The used Oligonucleotide primers were supplied from Metabion (Germany), as mentioned in Table (1).
PCR amplification was employed using 25 µL PCR reaction which was containing 12.5 µL of Emerald Amp Max PCR Master Mix (Emerald, Japan), 1 µL of each primer (20 pmol conc.), 4.5µL of PCR grade water and 6 µL of a template using a Biometra T3 thermal cycler. The PCR products were separated by the agarose gel electrophoresis using 1.5% agarose gel which stained with Ethidium bromide. The gel was photographed using a gel documentation system (Alpha Innotech, biometra).
The efficiency of the amplification was verified for positive field samples that might have mecA, aacA-aphD, tetK, mecA, sea, seb, sec, see, sed and tsst-1 genes which were previously examined in veterinary quality control reference laboratory for poultry production -Animal health research.
Results
Prevalence of S. aureus
There were 100 farms under examination for existence of S. aureus, only 21 farms (21%) were confirmed by S. aureus isolates. The 21 isolates were affirmed S. aureus as positive from the results of the slide catalase test, tube coagulase test and negative oxidase test.
Table 2: Antimicrobial susceptibility of S. aureus isolated from duck farms
| Antimicrobial |
Resistance % |
Intermediate % |
Sensitive % |
| Penicillin |
100 |
0 |
0 |
|
Ampicillin |
100 |
0 |
0 |
| Cefoxitin |
100 |
0 |
4 |
|
Oxacillin |
14.3 | 9.5 |
76.2 |
|
Gentamycin |
81 |
0 |
19 |
|
Kanamycin |
90.5 | 0 |
9.5 |
|
Ciprofloxacin |
4.8 | 19 |
76.2 |
|
Clindamycin |
28.6 | 23.8 |
47.6 |
|
Azithromycin |
28.6 | 23.8 |
47.6 |
|
Erythromycin |
23.8 | 66.7 |
9.5 |
|
Chloramphenicol |
19 | 0 |
81 |
| SXT | 14.3 | 0 |
85.7 |
|
Doxycycline |
14.3 | 28.6 |
57.1 |
|
Tetracycline |
85.7 | 4.8 |
9.5 |
|
AMC |
61.9 | 0 |
38.1 |
| CTX 30 | 9.5 | 57.1 |
33.4 |
|
Vancomycin |
0 | 0 |
21 |
|
Levofloxacin |
9.5 | 0 |
90.5 |
Antimicrobial Susceptibility Testing
All the 21 S. aureus isolates were tested against 18 antibiotics agents. The antimicrobial resistance profiles of the isolates to antimicrobial agents are shown in (Table 2). All MRSA isolates were resistant to penicillin, ampicillin and cefoxitin, in addition to high resistance rate to Kanamycin(90.5%), Tetracycline(85.7%) and Gentamycin(81%), while Ciprofloxacin showed the lowest resistance rate of 4.8%. In this work, the majority of isolates are classified as MDR-SA ones, as 19 isolates (90.5%) are resistant to three or more antimicrobial classes among different classes are well known as multidrug resistant (MDR), as shown in Figure (1), Table (2) and Table (4)
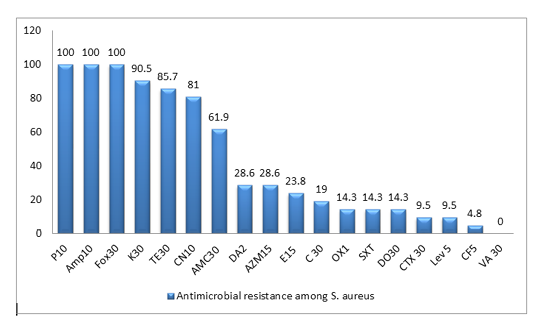
Figure 1: Antimicrobial resistance to 18 antimicrobials of S. aureus isolates from duck farms. The columns denote the percentages of resistant isolates for ducks). Penicillin-G (P 10); ampicillin (AMP 10);cefoxitin (FOX 30); Kanamycin (K 30); Tetracycline (T 30); gentamicin (CN 10); Amoxicillin + clavulanic acid (AMC 30); Clindamycin (DA2); Azithromycin (AZM 15); Erythromycin (E 15 ); Chloramphenicol (C 30 ); Oxacillin (OX1); Trimethoprim/sulfamethoxazole (SXT), Doxycycline (DO 30); cefotaxime (CTX 30); Levofloxacin (Lev 5); Ciprofloxacin (CF5 ); vancomycin (VA 30 )
Molecular Characteristics of S. aureus Isolates
Detection of presence of mecA and mecC genes:
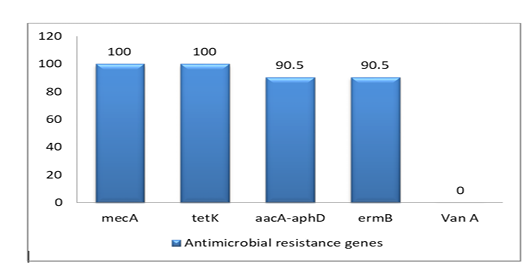
PCR has confirmed the presence of mecA gene in all S. aureus isolates (as 100%) as well as the absence of mecC gene.
Detection of Virulence-Associated Genes
Additionally, the PCR results for five classical enterotoxins (A–E) and toxic shock syndrome toxin 1(tsst-1) have revealed the MRSA isolates don’t own sea, seb, sec, and see genes and. On contrary for sed gene, only four isolates have had it.
Table 3: Correlation between phenotypic resistance and detection of resistance-associated genes in isolates
| Antimicrobial classes | Target genes | Phenotypic resistance | Gene detection |
|
Betalactam |
mecA |
21 | 21 (100%) |
|
Gentamicin |
aacA-aphD |
17 | 19(90.5%) |
|
Erythromycin |
ermB |
5 | 19 (90.5%) |
|
Tetracycline |
tetK |
18 | 21 (100%) |
|
Vancomycin |
VanA | 0 |
0 |
Table 4: Relationship between phenotypic antimicrobial resistance and detection of resistance genes and enterotoxin genes in MRSA isolated from ducks
| Strain | Phenotypic antibiotic resistance | Genotypic antibiotic resistance |
Type of se gene |
| 1 |
P10 , AMP10, FOX30, CN10, K30, TE30, AMC30 |
mecA, aacA-aphD, ermB, tetK |
|
| 2 |
P10 , AMP10, FOX30 ,CN10, K30, TE30, AMC30 |
mecA, aacA-aphD, tetK, mecA |
|
| 3 |
P10 , AMP10, FOX30,CN10, K30, TE30, AMC30 |
mecA, aacA-aphD, ermB, tetK |
|
| 4 |
P10 , AMP10, FOX30, OX1,CN10, K30, TE30, AMC30 |
mecA, aacA-aphD, ermB, tetK |
|
| 5 |
P10 , AMP10, FOX30,CN10 , K30, TE30, LEV5 |
mecA, aacA-aphD, ermB, tetK |
sed |
| 6 |
P10 , AMP10, FOX30, DA2, AZM15 ,E15, TE30 |
mecA, ermB, tetK |
|
| 7 |
P10 , AMP10, FOX30, OX1, AMC30 |
mecA, aacA-aphD, ermB, tetK |
|
| 8 |
P10 , AMP10, FOX30,CN10, K30, AZM15 , C30, AMC30 |
mecA, aacA-aphD, ermB, tetK |
sed |
| 9 |
P10 , AMP10, FOX30,CN10, K30, AZM15, TE30 |
mecA, aacA-aphD, ermB, tetK |
|
| 10 |
P10 , AMP10, FOX30,CN10, K30, TE30, CTX30 |
mecA, aacA-aphD, ermB, tetK |
|
| 11 |
P10 , AMP10, FOX30,CN10, K30, DA2,E15, SXT, DO30, , TE30, AMC30 |
mecA, aacA-aphD, ermB, tetK |
|
| 12 |
P10 , AMP10, FOX30,CN10, K30, TE30, AMC30 |
mecA, aacA-aphD, ermB, tetK |
|
| 13 |
P10 , AMP10, FOX30, OX1,CN10, K30 ,DA2,E15, SXT, , TE30 |
mecA, aacA-aphD, ermB, tetK |
|
| 14 |
P10 , AMP10, FOX30,CN10, K30, C30, DO30, TE30 |
mecA, aacA-aphD, ermB, tetK |
|
| 15 |
P10 , AMP10, FOX30,CN10, K30, TE30, AMC30 |
mecA, aacA-aphD, ermB, tetK |
|
| 16 |
P10 , AMP10, FOX30,CN10, K30, DA2, AZM15,E15, TE30 |
mecA, aacA-aphD, tetK |
|
| 17 |
P10 , AMP10, FOX30,CN10, K30, TE30, AMC30 |
mecA, ermB, tetK |
|
| 18 |
P10 , AMP10, FOX30,CN10, K30, DA2, AZM15, C30, TE30, AMC30 |
mecA, aacA-aphD, ermB, tetK |
sed |
| 19 |
P10 , AMP10, FOX30,CN10, K30, TE30 |
mecA, aacA-aphD, ermB, tetK |
|
| 20 |
P10 , AMP10, FOX30, K30, CIP 5, DA2, AZM15,E15 , C30, SXT , DO30, TE30 , AMC30, CTX30, LEV5 |
mecA, aacA-aphD, ermB, tetK |
Sed |
| 21 |
P10 , AMP10, FOX30, K30 |
mecA, aacA-aphD, ermB,, tetK |
Determination of antimicrobial Resistance Genes
The prevalence of 5 antimicrobial resistance genes testing was done for the isolates. The highest resistant gene were mecA and tetK (100%) to betalactam and tetracycline, whereas aacA-aphD and ermB genes were (90.5%) only. However, the VanA gene was negative as shown in Figure (2), Table (3) and Table (4)
Discussion
Many infections are caused basically by S. aureus whatever its zoonotic importance (Voss et al.,2005). Therefore, there are many reports about incidences of S. aureus and methicillin resistance in different poultry spp. such as chicken (Dweba et al., 2019) , turkey (El-Adawy et al., 2016) , and food products (Achek et al., 2018). There is lack or report about the incidence of S. aureus and methicillin resistance in ducks, especially in Egypt (Eid et al., 2019).
In this study, the prevalence of S. aureus results in ducks was (21%), In comparison to previous studies, this prevalence of S. aureus is higher than prevalence rates for 100 ducks in Egypt (12.2%) (Eid et al., 2019), Dutch duck farms (10%) (Van Duijkeren et al., 2016), and retail duck (7.2%) (Wang et al., 2017). However, the prevalence of S. aureus for this study is still lower than reported results for ducks in South African livestock (40%) (Dweba et al., 2019). ).The incidence of MRSA in duck farms represents a great concern to protect consumers. These results encourage us to assess the risk of any health hazards which may happen and become more curious to know the its possibility to induce infections , so this study showed important information about MRSA from duck farms based on phenotypic and genotyping characterization and detection of enterotoxin genes and some antimicrobial resistance determinants.
Moreover, the antimicrobial resistance is one of the most global threats to human causing severe public health diseases (Wang et al., 2017). Moreover, WHO is doing its supreme effort to support health project of control antibiotic resistance in humans and veterinary sector by cooperation with other organizations as the Food and Agriculture Organization of the United Nations (FAO) and the World Organization for Animal Health (OIE) (WHO, 2020). Especially for diseases have ability to be transmitted from food producing animals throughout food chain. Uncontrolled usage and intake abuse of antibiotics for either human or animals without physician or veterinarian advice will have been bad consequence in dissemination of antibiotic resistance (WHO, 2020). Hence, Staphylococcal infections are usually treated by excessive usage of penicillin and tetracycline. The misusage of those antibiotics leads to increase antimicrobial resistance (Nemati et al., 2008), as similar as to our antimicrobial resistance data in this current study.
It is found that the Cefoxitin is more accurate and is better than oxacillin in detection and identification of methicillin resistance. This regarding to Cefoxitin has high sensitivity, specificity and a higher effect on penicillin-binding protein 2a (PBP2a) (Marek et al., 2018) rather than oxacillin (CLSI, 2015).
In the present work, all MRSA isolates are having resistance to penicillin, ampicillin. There is agreement with previous reports for isolated MRSA from ducks in Egypt (Eid et al., 2019). Moreover, all resistant isolates to penicillin and ampicillin has recorded mecA gene which is the principle inducer for methicillin resistance (Marek et al., 2018).
In this study, 23.8% of S. aureus isolates were resistant to erythromycin, while 14.3% were resistant to SXT. Thus, S. aureus isolates were less resistant to SXT than erythromycin, but both are pretty less than reported for ducks in Egypt before (80% for each one) (Eid et al., 2019).
On other hand, the resistance of S. aureus against gentamycin (81%) was higher than what was reported previously in Egyptian duck farms (20%) (Eid et al., 2019), ducklings (26.7%) (Farghaly et al., 2015), and from duck and turkey farms in Netherlands (52.5%) (Van Duijkeren et al., 2016).
The isolated MRSA from ducks has shown 100% susceptibility to vancomycin in good agreement with previous results from Netherlands farms (Van Duijkeren et al., 2016).
Additionally, the antimicrobial resistance rates in this study for clindamycin and erythromycin (28.6%, & 23.8%) as shown in Table (2) respectively were lower than those of MRSA isolates from duck and turkey that reported in Netherlands farms (60% each). Nevertheless, the ciprofloxacin has higher resistance rate (52.5%) than reported rates in our work (4.8%) (Van Duijkeren et al., 2016).
In this study, 90.5% of S. aureus isolates exhibited multi-drug resistance as stated by others who are relating to poultry isolates (Dweba et al., 2019; El-Adawy et al., 2016). These MDR isolates are serious threats to human who is with direct contact with ducks in the infected farms (Van Duijkeren et al., 2016). Furthermore, the majority of MRSA isolates exhibit MDR and have high percentage of resistance to tetracycline and gentamycin as previously mentioned (Marek et al., 2018; Lyon and Skurray, 1987). Moreover as shown in Table (4), all of these isolates are resistant to penicillin which is mediated by penicillinase and all of them having mecA gene (Leonard and Markey, 2008).
For these reasons, the mecC and mecA genes are responsible for detection of beta-lactam resistance (El-Adawy et al., 2016). The mecC gene in MRSA is proved by numerous studies to be of zoonotic importance in livestock animals (Dweba et al., 2019). In this study, All MRSA isolates do not harbor mecC gene although mecA gene was detected in all isolates which is in agreement with previous study (Van Duijkeren et al., 2016).
Moreover, resistance of staphylococci to aminoglycoside (such as gentamicin, kanamycin and tobramycin) is mediated by aacA-aphD gene (Wendlandt et al., 2013; Achek et al., 2018; ). Thus, several studies have mentioned the prevalence of aminoglycoside and methicillin resistance relationship (Choi et al., 2003), with good agreement to our study as well. This relation can be explained as a result of neighboring positions of mecA gene and aminoglycoside resistance genes (Choi et al., 2003)
Although, all MRSA isolated from duck in China harbored aminoglycoside resistance gene aacA-aphD (Cao et al., 2016), whereas in this study only 90.5% of ducks MRSA isolated could be detected.
Moreover, MRSA from different origins as pigs, cattle, chickens and ducks commonly carried tet(K) and/or tet(L) (Wendlandt et al. 2013; Schwarz et al., 1998) . All MRSA isolates (100%) in this study also carry tetracycline resistance gene tetK as well as MRSA isolates of duck in China (Cao et al., 2016). On contrary, lower percentage of tetK gene from avian species (chicken, duck and wild birds in in South Africa (42.5%) was reported (Dweba et al., 2019).
However, there are several genes responsible for the resistance to macrolides, ermB is one of those genes. The ermB gene was found in 90.5% of isolates too. In spite of, the resistance due to ermB hasn’t been studied in ducks before, the resistance of ermB due to its occurrence in coagulase positive and coagulase-negative staph strains in poultry is 8.3% in USA farms (Nawaz et al., 2000), while it was 50% in turkey (El-Adawy et al., 2016).
Although, it is well known that vancomycin is the first choice drug for treatment of MRSA in human (Saha et al., 2008), but in 2002, the CDC (Centers for Disease Control and Prevention) declared a first case of S. aureus resistant to methicillin and vancomycin (CDC, 2002). Therefore, a continuous monitoring for vancomycin resistance is required.
Moreover, VanA gene associated with vancomycin resistance wasn’t detected in S. aureus isolates in this work. This result was in good agreement with the result reported by Ahmed et al (Ahmed et al., 2020). Recently, a new report has identified a first record of VanA gene where its detection came from camels’ meat in Egypt (Al-Amery et al., 2019).
The heat stable staphylococcal enterotoxins (SEs) are the main reason of food poisoning over the worldwide (Le Loir et al., 2003) . Thus, (Chao et al., 2015) reported that sea, seb, sec, and sed genes were not found except one strain harbored see gene in ducks, whereas the presence of sed gene was only found in four isolates in this work. Furthermore, all other classical enterotoxin genes and (tsst-1) weren’t detected in the current work which is similar to results recorded in Whiteface whistling ducks in Germany (Feßler et al., 2018).
Conclusion
The study of Methicillin-resistant S. aureus (MRSA) from ducks was conducted to acquire more information for hidden problem solving in duck farms in Egypt. Although S. aureus has low significance as a problem in ducks, but it is a real source for antimicrobial resistance spreading in surroundings and in vicinity of duck farms. MRSA has proved influences on public health of human and on the environment causing infection through livestock animals, so we need more investigation and surveillance studies were needed on the dissemination of MRSA.
Conflict of interest
The authors declare that there is no conflict of interest for this publication.
Authors’ contributions
Abdel Rahman: Methodology (Bacterial Isolation, Antimicrobial Susceptibility Tests), writing- reviewing and editing of manuscript. Amer: Methodology (Molecular Identification of S. Aureus Isolates Using Polymerase Chain Reaction Assay) and approved the final manuscript.
References