Advances in Animal and Veterinary Sciences
Research Article
Phenotypic and Genotypic Characterization of Salmonella enterica Serovars Isolated from Imported Poultry
Heba Badr*, Heba Roshdy, Hend K. Sorour, Mona A.A. AbdelRahman, Ahmed M. Erfan, Noha Salem, Soad A.Nasef, Wafaa M. Hassan, May F.Abdelaty
Reference Laboratory for Veterinary Quality Control on Poultry production, Animal Health Research Institute, Agricultural Research Center (ARC). Nadi El-Seid Street, Dokki P.O. Box 246, Giza 12618, Egypt.
Abstract | Monitoring of imported 1-day-old poultry is mandated in Egypt to prevent the possible introduction of new Salmonella serovars into the country’s poultry industry. Such serovars are considered to be a major public health threat. We examined 391 imported poultry flocks for the presence of salmonellae (231 duckling, 84 chick, and 76 turkey poult), serotyped all isolated salmonellae, and performed antimicrobial susceptibility testing. Molecular profiles were also constructed based on results of conventional polymerase chain reaction assays to detect virulence genes (stn, avrA, and sopB) and antibiotic resistance genes (blaTEM, tetA(A), and qnrS) in the Salmonella isolates. Thirty Salmonella strains were isolated from the 391 samples (7.7%). By poultry type, salmonellae were isolated from 21 of 231 (9.1%) duckling samples, 6 of 84 (7.1%) of chick samples, and 3 of 76 (3.9%) turkey poult samples. Serotyping of the isolates identified 16 different serovars: S. Enteritidis, S. Typhimurium, S. Sinstorf, S. Muenster, S. Vejle, S. Cuckmere, S. Indiana, S. Infantis, S. Koenigstuhl, S. Macallen, S. Nchanga, S. Neftenbach, S. Newlands, S. Nigeria, S. Nyborg, and S. Regent. The isolates showed variable degrees of antibiotic resistance across species. All tested Salmonella strains harbored the virulence and antibiotic resistance genes, with the exception of qnrS, which was found in only 50% of the isolates. In conclusion, examination of imported poultry is a critical point of control to prevent poultry from becoming a reservoir for human health hazards, including salmonellae with antimicrobial resistance phenotypes.
Keywords | Imported poultry, Salmonella serovars, sensitivity, virulence genes, antibiotic genes.
Received | February 03, 2021; Accepted | February 10, 2021; Published | May 01, 2021
*Correspondence | Heba Badr, Reference Laboratory for Veterinary Quality Control on Poultry production, Animal Health Research Institute, Agricultural Research Center (ARC). Nadi El-Seid Street, Dokki P.O. Box 246, Giza 12618, Egypt; Email: drheba_badr@yahoo.com
Citation | Badr H, Roshdy H, Sorour HK, AbdelRahman MAA, Erfan AM, Salem N, Nasef SA, Hassan WM, Abdelaty MF (2021). Phenotypic and genotypic characterization of salmonella enterica serovars isolated from imported poultry. Adv. Anim. Vet. Sci. 9(6): 823-834.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.6.823.834
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Badr et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
In addition to its role in food-borne illness all over the world, Salmonella is an important bacterial pathogen in poultry (Osman et al., 2010; Foley et al., 2011). Several serovars have been isolated from both poultry and humans, and poultry can transmit Salmonella to humans (Osman et al., 2010; European Food Safety Authority [EFSA], 2019).
The international trade of poultry is one of the factors affecting the spread of salmonellosis (EFSA, 2019) by acting as a portable reservoir for Salmonella (Osman et al., 2010), transmitting infection through the food production chain (Velhner et al., 2018), and vertically through the infected parents (Foley et al., 2011; Osman et al., 2014b), by transmission of multidrug-resistant (MDR) Salmonella in the Mediterranean region (Le Hello et al., 2013).
EFSA (2019) classified salmonellosis as the second human zoonotic disease underlying gastrointestinal illness and (Ezzeldeen et al., 2013) mentioned that the salmonellosis can cause multi-organ systemic infection. In the European Union (EU), salmonellae are estimated to cause almost 92,000 illnesses annually (EFSA, 2019). These illnesses are primarily caused by two serovars, Salmonella Enteritidis and Salmonella Typhimurium (Sodagari et al., 2019). However, non-typhoidal Salmonella (NTS) can also cause mild to severe illness and sometimes act as a means of transmission of antibiotic resistance to other Salmonella importance in poultry and human health (Osman et al., 2014b; Sodagari et al., 2019).
Uncontrolled and indiscriminate use of antibiotics for growth promotion or prophylaxis, especially in low- and middle-income countries, can lead to the emergence of antibiotic resistance to fluoroquinolones and extended-spectrum beta-lactam antibiotics in the poultry sector (Badr et al., 2015; Velhner et al., 2018). MDR salmonellae are significant for both human and animal health because they can lead to illness that is unresponsive to antibiotic treatment (Chen et al., 2013).
Antibiotic resistance is transmitted between bacteria through mobile genetic elements such as plasmids, transposons, and integrons. Such transmission results in healthy animals becoming carriers for antibiotic-resistant bacteria, which may subsequently cause human infections that are difficult to treat (Mthembu et al., 2019).
Bacterial self-defense against antibiotics involves developing modifications such as decreased cell permeability, alteration or replacement of the target, and enzyme inactivation that serve as mechanisms enabling antibiotic resistance (Frye and Jackson, 2013). For example, bacteria that are resistant to beta-lactam antibiotics produce beta-lactamase, an enzyme that can destroy the beta-lactam ring and thus inactivate the antibiotics (Mthembu et al., 2019). In salmonellae, many extended-spectrum beta-lactamases (ESBLs) exist, with tem, shv, and ctx-M genes being the most effective and frequent encoding genes (Jin and Ling, 2006).
In gram-negative bacteria, tetA is the most frequent antibiotic-resistance gene, and it may be present on either chromosomes or plasmids (Pezella et al., 2004). This gene enables tetracycline resistance in salmonellae that infect humans and animals (Threlfall, 2002). Qnr proteins are a group of penta peptide repeat proteins that allow bacteria to evade the effects of quinolones and develop resistance to these antibiotics. Qnr genes prevent or limit ciprofloxacin from disabling bacterial DNA gyrase and topoisomerase IV, enzymes that are necessary for bacterial DNA synthesis and thus growth (Tran and Jacoby, 2002).
Various virulence genes are important for Salmonella pathogenesis, such as sopB and avrA, which encode T3SS proteins that are involved in the Salmonella-induced inflammatory response and have anti-apoptotic roles. Together, the two proteins delay apoptosis of epithelial cells, which aids salmonellae in avoiding host adaptive immune response and in constructing a stable intracellular niche (Wu et al., 2011). Salmonella enterotoxin (stn) is an important virulence factor that causes diarrhea (Chopra et al., 1999). Some authors have reported that stn is not a virulence factor because they did not find any decrease in virulence phenotypes when the stn gene was deleted in tested mutant (Nakano et al., 2012). In contrast, earlier research (Chopra et al., 1999) found that inactivation of the stn gene reduced the ability of Salmonella Typhimurium ability to induce intestinal fluid accumulation.
The present study was conducted to determine the presence of salmonellae in chicks, ducklings, and poults being imported into Egypt. The isolates were studied with regard to their phenotypic characteristics and identified by serovar. In addition, their antimicrobial resistance profiles and genotypic profiles were defined based on important virulence genes (stn, avrA, and sopB) and antibiotic resistance genes (blaTEM, tetA(A), and qnrS).
Materials and Methods
Ethics approval
Birds were handled in accordance with the regulations for collecting samples from live animals, and this study was approved by the animal care committee of the Animal Health Research Institute.
Sampling
We obtained 391 samples from imported poultry flocks (231 ducklings, 84 chicks, and 76 turkey poults) from January to December 2019, by collecting the paper-lined the boxes used to transport the birds. Each box contained 30 young birds. The collected samples represented the pooled meconium samples from these 30 birds/box.
Isolation and characterization of salmonellae
Salmonella spp. were isolated and identified according to ISO 6579-1: 2017. The paper -lined the box (average 15-20gm) was placed in pre-enrichment medium 1:10 dilution (buffered peptone water; Oxoid, UK), which was subsequently incubated at 37°C for 16–18 h. Next, 0.1 mL of the pre-enrichment medium was transferred to modified semisolid Rappaport-Vassiliadis medium (MSRV, LabM, UK) and incubated at 41.5°C for 24 h. In addition, 1 mL of the pre-enrichment medium was transferred to MKTTn broth (LabM, UK) and incubated aerobically 37°C for 24 h, and then streaked onto XLD (LabM, UK) and SS (Oxoid, UK) agar plates, which were subsequently incubated at 37°C for 24 h aerobically. Selected colonies were then identified by biochemical tests (urea agar, triple sugar iron, and lysin iron) (Oxoid, UK).
Serotyping of Salmonella isolates
Next, the isolated Salmonella species were serotyped according to ISO 6579-3: 2014, and the serotypes were classified according to the Kauffman–White scheme (Grimont and Weill, 2007) using Salmonella antiserum (Sifin Co., Japan).
Antimicrobial susceptibility test
The antimicrobial sensitivity of each Salmonella strain was then tested. An antibiogram of the Salmonella isolates was created based on the results of disc-diffusion tests, conducted according to Koneman et al. (1997), using 13 antibiotics (ampicillin, apramycin, cefotaxime, ciprofloxacin, clindamycin, colistin sulphate, levofloxacin, lincomycin, nalidixic acid, norfloxacin, streptomycin, tetracycline, and trimethoprim-sulfamethoxazole; Oxoid). The results were interpreted according to CLSI/NCCLS (2017).
Molecular identification
A conventional polymerase chain reaction (PCR) assay was used to detect virulence and antibiotic-resistance genes in Salmonella isolates. DNA extraction was performed using the QIAamp DNA Mini kit (Qiagen, Germany, GmbH), with modifications of the manufacturer’s recommendations. Briefly, 200 µL of the sample suspension was incubated with 10 μL of proteinase K and 200 μL of lysis buffer at 56°C for 10 min. After incubation, 200 μL of 100% ethanol was added to the lysate. The sample was then washed and centrifuged following the manufacturer’s recommendations. Nucleic acid was eluted with 100 μL of elution buffer provided in the kit. The oligonucleotide primers used were supplied from metabion international AG (Germany) and are listed in Table 1. Each 25-μL reaction mixture contained 12.5 μL of EmeraldAmp Max PCR Master Mix (Takara, Japan), 1 μL of each primer (20 pmol concentrations), 5.5 μL of water, and 5 μL of DNA template. The reaction was performed in an Applied Biosystems 2720 thermal cycler.
The PCR products were separated by electrophoresis on 1.5% agarose gel (AppliChem, GmbH, Germany) in 1× TBE buffer at room temperature using gradients of 5 V/cm. For gel analysis, 20 μL of conventional PCR products were loaded into each gel slot. GeneRuler 100 bp DNA Ladder (Fermentas, Thermo, Germany) was used to determine the fragment sizes. The gel was photographed with a gel documentation system (Alpha Innotech, Biometra, Germany), and the data were analyzed through computer software.
Results
Prevalence of Salmonella
Among the 391 imported poultry flock samples (231 duckling, 84 chick, and 76 turkey poult) collected, 30 (7.7%) were positive for Salmonella. Based on species, 21 of 231 (9.1%) duckling samples, 6 of 84 (7.1%) chick samples, and 3 of 76 (3.9%) poult samples yielded Salmonella isolates as shown in Table (2).
Table 2: Number of Salmonella isolated from different imported poultry samples
| Samples | Number of tested samples | Positive Salmonella isolation * | |
| No. | (%)* | ||
| Duckling | 231 | 21 | (9.1%) |
| Chicks | 84 | 6 | (7.1%) |
| Poults | 76 | 3 | (3.9%) |
| Total | 391 | 30 | (7.7%) |
(%)*: Number of positive related to number of tested samples.
Serotyping of the Salmonella strains isolated from the three different bird species revealed a broad range of serovars. Poults had three different serotypes (S. Enteritidis, S. Muenster, and S. Cuckmere), and chicks had two strains for both S. Typhimurium and S. Vejle and one each for S. Enteritidis and S. Sinstorf. Ducklings had four S. Enteritidis strains, three S. Vejle strains, and two S. Nyborg strains, as well as one strain for each of the following serotypes: S. Typhimurium, S. Sinstorf, S. Muenster, S. Indiana, S. Infantis, S. Koenigstuhl, S. Macallen, S. Nchanga, S. Neftenbach, S. Newlands, S. Nigeria, S. Nyborg, and S. Regent as shown in Table (3).
Antimicrobial susceptibility
All strains showed variable degrees of antibiotic resistance as shown in Table 4. The most frequent resistance phenotypes involved clindamycin and lincomycin, with all species being resistant to these antibiotics. Nalidixic acid resistance was found in 57.2% of the Salmonella isolates from ducklings and 16.7% of those from chicks. A High level of trimethoprim-sulfamethoxazole resistance was found in isolates from chicks and poults, with 50% and 66.7% of isolates being resistant, respectively. Resistance to ampicillin was found for 38% of isolates from ducklings and 33.3% of those from both chicks and poults. Tetracycline resistance was observed for 38.1% of isolates from ducklings and 33.3% of those from chicks. Streptomycin resistance was associated with 38.1% of isolates from ducklings, 16.7% from chicks, and 66.7% from poults. Lower levels of resistance were found for cefotaxime, with 33.3% of duckling isolates showing resistance, and for ciprofloxacin, with 28.6% of duckling isolates and 33.3% of both chick and
Table 1: Primers sequences, target genes, amplicon sizes and cycling conditions.
| Target genes | Primers sequences | Amplified segment (bp) | Annealing | Reference |
| stn | TTGTGTCGCTATCACTGGCAA CC | 617 |
59˚C 40 sec. |
Murugkar et al., 2003 |
| ATTCGTAACCCGCTCTCG TCC | ||||
| avrA | CCT GTA TTG TTG AGC GTC TGG | 422 |
58˚C 40 sec. |
Huehn et al. 2010 |
| AGA AGA GCT TCG TTG AAT GTC C | ||||
| sopB | TCAGAAGRCGTCTAACCACTC | 517 |
58˚C 40 sec. |
Thung et al., 2018 |
| TACCGTCCTCATGCACACTC | ||||
| qnrS | ACGACATTCGTCAACTGCAA | 417 |
55˚C 40sec. |
Robicsek et al., 2006 |
| TAAATTGGCACCCTGTAGGC | ||||
| tetA (A) | GGTTCACTCGAACGACGTCA | 576 |
50˚C 40 sec. |
Randall et al. 2004 |
| CTGTCCGACAAGTTGCATGA | ||||
| bla TEM | ATCAGCAATAAACCAGC | 516 |
54˚C 40 sec. |
Colom et al., 2003 |
| CCCCGAAGAACGTTTTC |
Thermo cycler program: Primary denaturation 94˚C/5 min. then Amplification (35 cycles) {Secondary denaturation 94˚C/ 30 sec., Annealing differ from each primer, Extension 72˚C/ 45 sec.} then final extension72˚C/ 10 min.
Table 3: Salmonella serovars with antigenic formula in different samples
| Serotypes | Ducklings (21) | Chicks(6) | Poults (3) |
|
S. Enteritidis "1,9,12;g,m;--" |
4 | 1 | 1 |
|
S.Typhimurium "1,4,5,12;i;1,2" |
1 | 2 | - |
|
S. Sinstorf "3,10;l,v;1,5" |
1 | 1 | - |
|
S. Muenster "3,10;e,h;1,5" |
1 | - | 1 |
|
S. Vejle "3,10;e,h;1,2" |
3 | 2 |
- |
|
S. Cuckmere "3,10;i;1,2" |
- | - | 1 |
|
S. Indiana "1,4,12;z;1,7" |
1 | - | - |
|
S. Infantis "6,7,14;r;1,5" |
1 | - | - |
|
S. Koenigstuhl "1,4,[5],12;z;e,n,z15" |
1 | - | - |
|
S. Macallen "3,10;z36,-" |
1 | - | - |
|
S. Nchanga "3,10,15;l,v;1,2" |
1 | - | - |
|
S. Neftenbach "4,12;z;e,n,x" |
1 | - | - |
|
S. Newlands "3,10;e,h;e,n,x" |
1 | - | - |
|
S. Nigeria "6,7;r;1,6" |
1 | - | - |
|
S. Nyborg ""3,10;e,h;1,7" |
2 | - | - |
|
S. Regent "3,10;f,g,s;1,6" |
1 | - |
- |
Table 4: Interpretation of antibiotic sensitivity test for Salmonella isolates
| Antibiotics | Ducklings N=21(%) | Chicks N=6 (%) | Poults N=3 (%) | ||||||
| R | I | S | R | I | S | R | I | S | |
|
AMP (10 µg) |
8 (38) | - | 13 (62) | 2 (33.3) | - | 4 (66.7) | 1 (33.3) | - | 2 (66.7) |
|
APR (15 µg |
4 (19) | 9 (43) | 8 (38) | - | 1 (16.7) | 5 (83.3) | - | 2 (66.7) | 1 (33.3) |
|
CTX (30 µg) |
7 (33.3) | 5 (23.7) | 9 (43) | - | 2 (33.3) | 4 (66.7) | - | 1 (33.3) | 2 (66.7) |
|
CIP (5 µg) |
6 (28.6) | 14 (66.6) | 1 (4.8) | 2 (33.3) | 3 (50) | 1 (16.7) | 1 (33.3) | 2 (66.7) | - |
|
DA (2 µg) |
21 (100) | - | - | 6 (100) | - | - | 3 (100) | - | - |
|
CT (10 µg) |
1 (4.8) | - | 20 (95.2) | - | - | 6 (100) | 1 (33.3) | - | 2 (66.7) |
|
LEV (5 µg) |
3 (14.3) | 2 (9.5) | 16 (76.2) | - | - | 6 (100) | - | - | 3 (100) |
|
MY (10 µg) |
21 (100) | - | - | 6 (100) | - | - | 3 (100) | - | - |
|
NA (30 µg) |
12 (57.2) | 2 (9.5) | 7 (33.3) | 1 (16.7) | 1 (16.7) | 4 (66.7) | - | 1 (33.3) | 2 (66.7) |
|
NOR (10 µg) |
1 (4.8) | 2 (9.5) | 18 (85.7) | - | 1 (16.7) | 5 (83.3) | - | - | 3 (100) |
|
S (10 µg) |
8 (38.1) | 11 (52.4) | 2 (9.5) | 1 (16.7) | 3(50) | 2 (33.3) | 2 (66.7) | 1 (33.3) | - |
|
TE (30 µg) |
8 (38.1) | 1 (4.8) | 12 (57.1) | 2 (33.3) | - | 4 (66.7) | - | - | 3 (100) |
|
SXT (1.25/23.75 µg) |
3 (14.3) | - | 18 (85.7) | 3 (50) | - | 3 (50) | 2 (66.7) | - |
1 (33.3) |
R: Resistance, I: Intermediate, S: Sensitive
AMP= Ampicillin, APR= Apramycin, CTX= Cefotaxime, CIP= Ciprofloxacin, DA= Clindamycin, CT= Colistin sulphate, LEV= Levofloxacin, MY= Lincomycin, NA= Nalidixic acid, NOR= Norfloxacin, S= Streptomycin, TE= Tetracycline, SXT= Trimethoprim-sulfamethoxazole.
Table 5: Identification of antibiotic resistance and virulence genes in Salmonella isolates
| Sample | blaTEM | tetA(A) | qnrS | stn | avrA |
sopB |
| Duckling (21 isolates) | 21/21 (100%) | 21/21 (100%) | 10/21 (47.6%) | 21/21 (100%) | 21/21 (100%) | 21/21 (100%) |
| Chicks (6 isolates) | 6/6 (100%) | 6/6 (100%) | 3/6 (50%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) |
| Poults (3 isolates) | 3/3 (100%) | 3/3 (100%) | 2/3 (66.6%) | 3/3 (100%) | 3/3 (100%) | 3/3 (100%) |
| Total | 30/30 (100%) | 30/30 (100%) | 15/30 (50%) | 30/30 (100%) | 30/30 (100%) |
30/30 (100%) |
Table 6: Virulence and antibiotic resistance genes and antibiotic resistance profiles of Salmonella Serovars:
| Code No. | Sample species | Salmonella Serovars | Antibiotic resistance genes | Virulence genes | Antibiotic resistance profile/ resistance (intermediate) |
| 1 | Duckling | Enteritidis |
blaTEM , tetA(A) |
stn, avrA, sopB | DA, MY (CTX, S, CIP) |
| 2 | Duckling | Macallen |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | CTX, DA, MY (S, APR, CIP) |
| 3 | Duckling | Enteritidis |
blaTEM ,tetA(A), qnrS |
stn, avrA, sopB | DA, MY (CTX, CIP) |
| 4 | Chicks | Sinstorf |
blaTEM , tetA(A) |
stn, avrA, sopB | DA, MY (CIP) |
| 5 | Duckling | Typhymurium |
blaTEM , tetA(A) |
stn, avrA, sopB | DA, MY, NA (S, CIP) |
| 6 | Chicks | Typhymurium |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | DA, SXT, MY (S, CIP) |
| 7 | Duckling | Regent |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | DA, APR, MY, NA (CTX, TE, S, CIP) |
| 8 | Duckling | Kenigstuhl |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | DA, MY, NA (S, CIP) |
| 9 | Poults | Enteritidis |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | DA, SXT, MY, CIP (S, APR, NA) |
| 10 | Duckling | Enteritidis |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | AMP, CTX, DA, TE, S, MY, NA, CIP (NOR, LEV) |
| 11 | Duckling |
Nyborg |
blaTEM , TetA(A) |
stn, avrA, sopB | S, CTX, DA, TE, S, MY, NA, CIP (LEV) |
| 12 | Chicks | Vejle |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | DA, MY, CIP (CTX, S, APR) |
| 13 | Poults | Cuckmere |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | AMP, DA, SXT, S, MY (APR, CIP) |
| 14 | Chicks | Vejle |
blaTEM , tetA(A) |
stn, avrA, sopB | DA, SXT, MY (S) |
| 15 | Duckling | Nchanga |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | CTX, DA, MY (S, APR, NA, CIP) |
| 16 | Duckling | Vejle |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | AMP, DA, NOR, SXT, TE, S, LEV, CT, APR, MY, NA, CIP |
| 17 | Duckling | Neftenbach |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | DA, S, APR, MY, NA (CIP) |
| 18 | Duckling | Sinstorf |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | DA, SXT, MY (S, APR) |
| 19 | Duckling | Muenster |
blaTEM , tetA(A) |
stn, avrA, sopB |
DA, MY (S, APR, CIP) |
| 20 | Duckling | Vejle |
blaTEM , tetA(A) |
stn, avrA, sopB | AMP, DA, TE, MY (APR, NA, CIP) |
| 21 | Duckling | Enteritidis |
blaTEM , tetA(A) |
stn, avrA, sopB | AMP, CTX, DAM TE, S, LEV, MY, NA, CIP (APR) |
| 22 | Poults | Muenster |
blaTEM , tetA(A) |
stn, avrA, sopB | DA, S, CT, MY (CTX, CIP) |
| 23 | Duckling | Newlands |
blaTEM , tetA(A) |
stn, avrA, sopB | AMP, CTX, DA, TE, S, MY (CIP) |
| 24 | Duckling | Vejle |
blaTEM , tetA(A) |
stn, avrA, sopB | DA, MY (S, CIP) |
| 25 | Duckling | Indiana |
blaTEM , tetA(A) |
stn, avrA, sopB | DA, MY, NA (CTX, S, CIP) |
| 26 | Chicks | Enteritidis |
blaTEM , tetA(A) |
stn, avrA, sopB | AMP, DA, TE, MY, NA, CIP (CTX, NOR) |
| 27 | Duckling | Infantis |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | AMP, CTX, DA, TE, S, LEV, APR, MY, NA, CIP (NOR) |
| 28 | Chicks | Typhymurium |
blaTEM , tetA(A), qnrS |
stn, avrA, sopB | AMP, DA, SXT, TE, S, MY (NA, CIP) |
| 29 | Duckling | Nyborg |
blaTEM , tetA(A) |
stn, avrA, sopB | AMP, DA, SXT, TE, S, MY, NA, CIP (CTX) |
| 30 | Duckling | Nigeria |
blaTEM , tetA(A) |
stn, avrA, sopB | DA, MY, NA (S, CIP) |
poult isolates showing resistance. Colistin sulphate resistance was detected in 33.3% of isolates from poults. On the other hands, each strain separately identified their antibiotic resistance on Table 5.
Molecular identification
To determine the virulence and antibiotic resistance profiles for all of the isolated Salmonella spp. (30 isolates), PCR was performed for the related genes. We found stn, avrA, and sopB virulence genes in all tested samples. With regard to antibiotic resistance, all tested strains carried blaTEM and tetA(A) genes, while the qnrS gene was reported in 50% of the isolates (Table 5) and Fig. (1, 2, 3, 4, 5 & 6).
On association relationship between species, resistance patterns, related antibiotic resistance genes and the virulence genes individually with its serotype as shown on Table (6); clarify the significance of each isolate and throw light about the multidrug resistance profile.
Discussion
Poultry plays an important role in Salmonella transmission because the animals serve as a reservoir. Although the birds may appear to be healthy, they intermittently shed Salmonella, and many reports have documented true- and/or pseudo-vertical transmission of Salmonella from chickens (EFSA, 2012). Examination of imported poultry flocks in the current study revealed that the incidence of Salmonella from 1-day-old poultry of different species was 7.7%, overall; by species, Salmonella was isolated from 9.1% of duckling samples, 7.1% of chick samples, and 3.9% of turkey poult samples. Each type of poultry carried multiple serotypes of Salmonella. Ducklings had the most serotypes (S. Enteritidis, S. Typhimurium, S. Sinstorf, S. Muenster, S. Vejle, S. Indiana, S. Infantis, S. Koenigstuhl, S. Macallen, S. Nchanga, S. Neftenbach, S. Newlands, S. Nigeria, S. Nyborg, and S. Regent), while chicks carried S. Enteritidis, S. Typhimurium, S. Sinstorf, and S. Vejle, and poults carried S. Enteritidis, S. Muenster, and S. Cuckmere. Jodas and Hafez (2002) reported similar results, isolating S. Enteritidis from 7 of 231 (3%) of meconium samples from poults. In addition, rates of 4.3% and 4% were reported by Abdallaha et al. (2015) and Osman et al. (2014b), respectively, for poults imported into Egypt, while Sorour et al. (2016) and Osman et al. (2010) isolated Salmonella from 11.3% (6/53) and 12.6% of imported poults to Egypt, respectively.
The prevalence of Salmonella in ducklings in our study was substantially similar to the 2.1% found by Wang et al. (2020) isolated from China and the 6.45% reported by Badr et al. (2015) for imported ducklings into Egypt; se
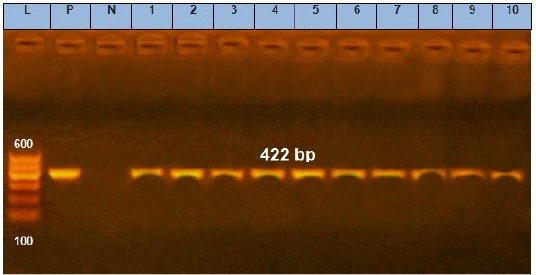
Figure 1: Agarose gel electrophoresis showing amplification of 422 bp fragments of avrA gene. Lane (1 to 10) shows the positive amplification of ten representing isolates. L: Ladder (100-600). P: Positive control and N: Negative control.
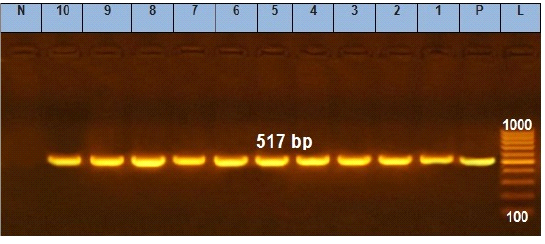
Figure 2: Agarose gel electrophoresis showing amplification of 517 bp fragments of sopB gene. Lane (1 to 10) shows the positive amplification of ten representing isolates. L: Ladder (100-1000). P: Positive control and N: Negative control.
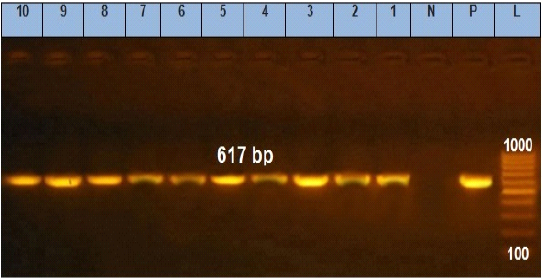
Figure 3: Agarose gel electrophoresis showing amplification of 617 bp fragments of stn gene. Lane (1 to 10) shows the positive amplification of ten representing isolates. L: Ladder (100-1000). P: Positive control and N: Negative control.
rotyped as S. Derby, S. Newport, S. Togo and S. Ball. Yang et al. (2019) reported a higher percentage of samples with Salmonella (12.2 %) from China, and Osman et al. (2010) and Osman et al. (2014c) similarly reported high percentages of 19.3% and 18.5% (25/135), respectively, among ducklings imported into Egypt. Ezzeldeen et al. (2013) documented a prevalence of 36% among duckling import
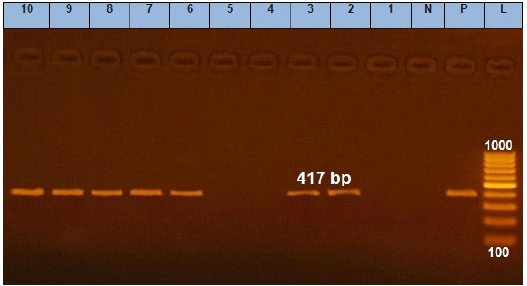
Figure 4: Agarose gel electrophoresis showing amplification of 417 bp fragments of qnrS gene. Lane (1 to 10) shows the amplification result of ten representing isolates. L: Ladder (100-1000). P: Positive control and N: Negative control.
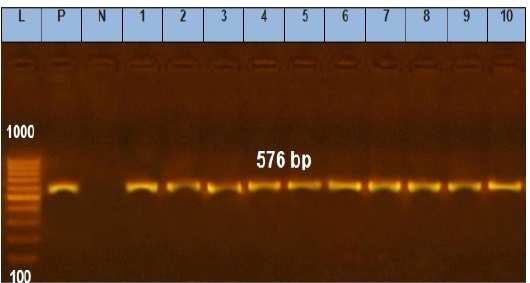
Figure 5: Agarose gel electrophoresis showing amplification of 576 bp fragments of tetA(A) gene. Lane (1 to 10) shows the positive amplification of ten representing isolates. L: Ladder (100-1000). P: Positive control and N: Negative control.
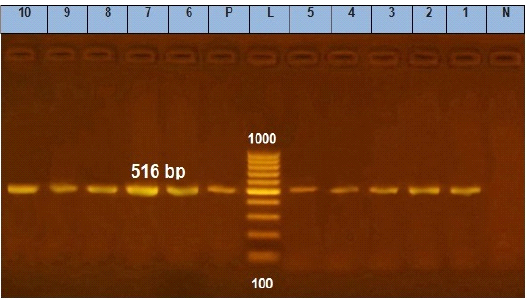
Figure 6: Agarose gel electrophoresis showing amplification of 516 bp fragments of blaTEM gene. Lane (1 to 10) shows the positive amplification of ten representing isolates. L: Ladder (100-1000). P: Positive control and N: Negative control.
ed into Egypt, and they identified the serotypes as S. Enteritidis, S. Kentucky, S. Newport, S. Brenderup, S. Tishiongwe, and S. Shubra. Habing et al. (2015) analyzed duckling samples into USA and showed that 30% (9/30) contained four serovars, including S. Typhimurium, S. Mbandaka, S. Infantis, and S. Lille, which were associated with outbreaks in 2013. In Brazil, Ribeiro et al. (2006) found 26 of 40 (65.0%) imported flocks were positive for Salmonella, which included the serovars S. Indiana, S. Newport, S. Saintpaul, S. Kottbus, S. Agona, S. London, S. Chester, and S. Kentucky. Wang et al. (2020) reported serovars S. Anatum, S. Apeyeme, and S. Montevideo.
Similar results were reported by Wang et al. (2020) who found a Salmonella prevalence of 4.2% (19/450) in 1-day-old chicks, Abdel Rahman et al. (2014) who reported 5.2% in imported chicks into Egypt, and Shehata et al. (2019) who demonstrated 7% prevalence in broiler chicks in Egypt. In contrast, Osman et al. (2014a) found a higher prevalence rate, with Salmonella isolated from 11.8% of imported chicks into Egypt in their study (13/110). The serotypes were S. Enteritidis, S. Typhimurium, S. Dublin, S. Shagoua, S. Hindmarch, and S. Inganda, with one isolate being untypeable. Habing et al. (2015) also found a high prevalence rate in their study that examined Salmonella from hatchling shipment boxes that contained chicks. That study showed a prevalence of 27.1% (44/162).
Sharma et al. (2018) isolated Salmonella from shipping boxes of hatchling poultry into U.S and identified multiple serovars, including S. Indiana, S. Muenster, S. Typhimurium, S. Enteritidis, S. Infantis, and 11 others. Shehata et al. (2019) isolated Salmonella from 7%, 12.5%, and 15% of samples for broilers, dead chicken embryos, and dead duck embryos; these isolated included S. Typhimurium, S. Enteritidis, S. Vejle, and S. Infantis, among others. In addition, Abdallaha et al. (2015) isolated Salmonella from chicks, ducklings, and poults into Egypt, and serotyping revealed 17 serovars, including S. Enteritidis, S. Newlands, and S. Nigeria.
Recently, antibiotic resistance has been increasing in Salmonella species isolated from animal origin with increasing emergence of drug-resistant strains. The common use of antibiotics to promote growth and to treat diseases in the poultry industry adds to the potential risk of the dissemination of MDR salmonellae (Abdallaha et al., 2015). In particular, antibiotic resistance of Salmonella associated with hatchlings is due to dipping eggs in an antibiotic solution or inoculation of hatching eggs with antimicrobial agents (Kabir, 2010).
In the present study, salmonellae isolated from ducklings showed high resistance to clindamycin and lincomycin (100%) but less resistance to nalidexic acid (57.2%) and ampicillin, streptomycin, and tetracycline (38%). The isolates had even less resistance to cefotaxime (33.3%) and ciprofloxacin (28.6%), followed by trimethoprim-sulfamethoxazole and levofloxacin (14.3%). Badr et al. (2015) reported isolates with high resistance to ciprofloxacin and nalidixic acid (reaching 75%), while Abdallaha et al. (2015) observed maximum resistance to trimethoprim (70.5%) followed by penicillin (41%), streptomycin (29.5%), nalidixic acid (23.5%), and ciprofloxacin (5.8%) in Salmonella strains isolated from ducklings, chicks, and poults.
In the current study, isolates from chicks showed high resistance to clindamycin and lincomycin 100%, then trimethoprim-sulfamethoxazole (50%) followed by tetracycline, ampicillin, and ciprofloxacin (33%). Similar results were reported for isolates from chicks by Abdel Rahman et al. (2014), who found resistance of 100%, 75%, 50%, 50%, and 50% to streptomycin, nalidixic acid, ciprofloxacin, tetracycline, and trimethoprim-sulfamethoxazole, respectively. Meanwhile, Osman et al. (2014a) documented resistance to lincomycin and susceptibility to ciprofloxacin and colistin sulfate in all Salmonella serotypes isolated from chicks, and Shehata et al. (2019) reported resistance to ampicillin, neomycin, trimethoprim-sulfamethoxazole, and streptomycin.
The results of this study showed that isolates from poults had high antimicrobial resistance to clindamycin and lincomycin (100%), followed by trimethoprim-sulfamethoxazole and streptomycin (66.7%) and colistin sulphate, ampicillin, and ciprofloxacin (33.3%). In comparison, Sorour et al. (2016) reported high resistance among Salmonella isolates turkey poults to lincomycin and ampicillin (100%), less resistance to ciprofloxacin and nalidixic acid (83.5%), and very low resistance to streptomycin (16.6%).
Wang et al. (2020) reported that the most frequent resistance phenotypes in samples from chicks and ducklings were for nalidixic acid (100%), streptomycin (92.7%), and ampicillin (92.7%), followed by tetracycline (40%) and ciprofloxacin (22.33%). Yildirim et al. (2011) found that Salmonella isolates were highly resistant to lincomycin, ampicillin, ciprofloxacin, and nalidixic acid. In contrast, Habing et al. (2015) reported that antimicrobial resistance was uncommon in Salmonella isolated from poultry hatchling; however, 19% (11/59) of the isolates were resistant to more than one class of antimicrobials.
With regard to virulence, some researchers (Wallis et al., 1999) have argued that the stn gene cannot be considered as a Salmonella virulence factor and it is not related to Salmonella enterotoxicity, while other research groups have reported that the stn gene is specific for all Salmonella serotypes (Lee et al., 2009). This finding highlights the stn gene as a reliable marker for Salmonella screening (Chopra et al., 1999). In the current study, the stn gene was detected in all tested samples, indicating that all isolates had the potential to produce the heat-labile exotoxin that is one of the main sharing agents in diarrhea (Van Asten and Van Dijk, 2005).
The 100% prevalence of sopB and avrA genes in the tested samples strengthened the virulence profiles of the isolated samples and was in agreement with different earlier research studies in poultry (Osman et al., 2014c; ElSheikh et al., 2019) and in humans (Astolfi-Ferreira et al., 2017).
Although, 100% of the tested isolates were positive for blaTEM gene by PCR, not all of them showed a resistance profile for one of beta-lactamases that inhibit antibiotics. This finding may have been related to poor expression of a functional TEM-1 enzyme due to promoter region mutation, or due to the presence of a mutant TEM enzyme that is inactive (Bradford et al., 2001). The discrepancy between the phenotypic resistance profile for quinolones and the qnrS gene PCR result is acceptable because quinolone resistance is mediated in three ways, including plasmid-mediated resistance genes (qnr genes), mutations in the quinolone resistance determining regions (QRDRs), and overexpression of efflux pumps mediated by qepA genes (Lunn et al., 2010). In this order, Egyptian regulations concerning the Salmonella in the imported live birds was condamination of the infected flocks with Salmonella specially S.Typhimurium and S.Enteritidis
Conclusion
Imported animals may circulate S. enterica serovars with the potential for multi-drug resistance and international spread, leading to a public health hazard. Isolation of different Salmonella serovars in this study suggested the introduction of new serotypes into Egypt and illustrated the ability of poultry to be a common reservoir for various Salmonella serovars in addition to the most dominant two (S. Enteritidis and S. Typhimurium). These results indicate the importance of applied risk assessment of Salmonella in Egypt and provide a guideline for control and continuous survey for identifying circulating Salmonella species in field.
Acknowledgments
We would like to thank the Reference Laboratory for Veterinary Quality Control on Poultry Production (RLQP) for providing the infrastructure for conducting the study. The authors did not receive any funds for this study.
Conflict of Interest
The authors declare that they have no conflict of interests.
authors contribution
HB designed the study. HB, HR, HKS, MAA, MFA and AME performed the research and drafted the manuscript. HB and AME analyzed the data. HB, HR, HKS, MAA, MFA and AME write, revised and finalized the manuscript for submission. All authors read and approved the final manuscript.
References





