Advances in Animal and Veterinary Sciences
Research Article
Advances in Animal and Veterinary Sciences 2 (2): 81 – 85Prevalence of Listeria spp. in Animals and Associated Environment
Abhay Raorane1, Swapnil Doijad1, Shilpa Katkar2, Ajay Pathak1, Krupali Poharkar1, Zunjar Dubal1, Sukhadeo Barbuddhe1*
- Indian Council of Agricultural Research Complex for Goa, Old Goa, Goa 403402 India
- Mumbai Veterinary College, Parel, Mumbai 400 012 India
*Corresponding author: barbuddhesb@yahoo.com
ARTICLE CITATION:
Raorane AV, Doijad S, Katkar S, Pathak A, Poharkar K, Dubal ZB, Barbuddhe SB (2014). Prevalence of listeria spp. in animals and associated environment. Adv. Anim. Vet. Sci. 2 (2): 81 – 85.
Received: 2013–12–21, Revised: 2013–12–30, Accepted: 2013–12–31
The electronic version of this article is the complete one and can be found online at
(
http://dx.doi.org/10.14737/journal.aavs/2014/2.2.81.85
)
which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
ABSTRACT
A total of 215 samples, comprising of both clinical (n=182) and farm environment (n=33), were screened to determine the prevalence of Listeria spp. in different parts of Konkan region, India. Out of these, 27 (12.55%) samples were found positive for Listeria spp. The isolates were further characterized using phenotypic assays (Hemolysis test, Christie Atkins Munch–Petersen test (CAMP) and growth on Agar Listeria acc. to Ottaviani & Agosti (ALOA)) and genotypic assays. The isolates were confirmed as Listeria monocytogenes 12(5.11%), L. ivanovii 2(0.93%), L. innocua 11(5.11%), L. seeligeri 2(0.93%) and L. welshimeri 1(0.46%). The L. monocytogenes isolates were recovered from clinical cases in sheep, goat and pig while one isolate was obtained from pig rearing environment. Serotyping of L. monocytogenes revealed 5 isolates to be of serotype 4b and 6 of serotype 1/2b. L. monocytogenes isolates were sensitive to ampicillin, doxycycline, ciprofloxacin, vancomycin and intermediate resistance towards chloramphenicol, penicillin and gentamicin. The study shows the prevalence of the L. monocytogenes in the clinical cases and associated farm environment of the Konkan region. However, further study is necessary to determine whether farm environment acts as potential reservoir transferring L. monocytogenes to animals.
INTRODUCTION
L. monocytogenes is Gram positive, facultative food borne pathogen of humans and animals (Dhama et al., 2013). The genus Listeria includes 10 species; L. monocytogenes, L. ivanovii, L. grayi, L. innocua, L. seeligeri, L. welshimeri, L. marthii, and L. rocourtiae, (Graves et al., 2010; Leclercq et al., 2010) including two recently identified species, L. fleischmannii, and L. weihenstephanensis (Halter et al., 2013; Bertsch et al., 2013). Out of these, L. monocytogenes and L. ivanovii are pathogenic to humans and animals (Guillet, 2010). The L. monocytogenes is widely distributed in the environment and has been isolated from a variety of sources including water, sludge, soil, plants, vegetation, food, food processing plants and infected humans and animals (Liu, 2008; Dhama et al., 2013).
The listeriosis is a rare but serious food–borne disease as it exhibits 20–30% mortality, 91% hospitalization and 50% neonatal death rates (Low and Donachie, 1997; Kathariou, 2002). In human, the early stage of infection by L. monocytogenes generally displays initial flu–like symptoms such as chilling, nausea, fever, gastroenteritis etc. Untreated cases may lead to septicemia, meningitis, encephalitis, abortion and occasionally death (Barbuddhe et al., 2008). Listeria infection in animal shows broad range of symptoms from asymptomatic infection to uncommon cutaneous lesions or various focal infections such as conjunctivitis, urethritis, endocarditis, and severe disturbance of the gait, followed by death. The L. monocytogenes is a well–recognized cause of mastitis, abortion, repeat breeding, infertility, encephalitis, and septicemia in cattle (Barbuddhe et al., 2008; Deb et al. 2013).
The infection by L. monocytogenes is generally transmitted through contaminated food (Farber and Peterkin, 1991). Generally industrially processed foods such as raw meat, fish, milk, milk–related products has been linked for the listeriosis (Rocourt, 2000; Lyytikäinen et al.,2006; Kvistholm Jensen et al., 2010; Goulet et al., 2012; Dhama et al., 2013)..
Heavy use of antibiotics as a growth promoter for farm animals and injudicious use of antibiotics accelerated evolution of bacteria towards antibiotic resistance. Since bacteria have the remarkable ability to develop resistance against antibiotics, bacterial species such as Listeria has also evolved towards multiple antibiotics resistance (Charpentier and Courvalin, 1999).
In India, the occurrence of listeriosis is poorly studied. Lack of awareness makes L. monocytogenes underdiagnosed therefore incidences in humans and animals are underestimated. Several researchers have explored incidence of listerial spp. in India from different sources (Jallewar et al., 2007; Yadav et al., 2011; Manoj et al., 1991; Dhanashree et al., 2003; Moharem et al., 2007; Kalorey et al., 2008). Also, L. monocytogenes cases have been reported sporadically in humans and animals (Mokta et al., 2010; Adhikary and Joshi, 2010). The present epidemiological data is insufficient and not conclusive (Barbuddhe et al., 2012). The objective of the study is to determine the prevalence of Listeria spp. in Konkan region of Maharashtra, India.
MATERIALS AND METHODS
Bacteria
Standard cultures of L. monocytogenes (MTCC 1143), Staphylococcus aureus (MTCC 1144), and Rhodococcus equi (MTCC 1135) were obtained from Microbial Type Culture Collection Center, Institute of Microbial Technology (IMTECH), Chandigarh, India.
Sample Collection
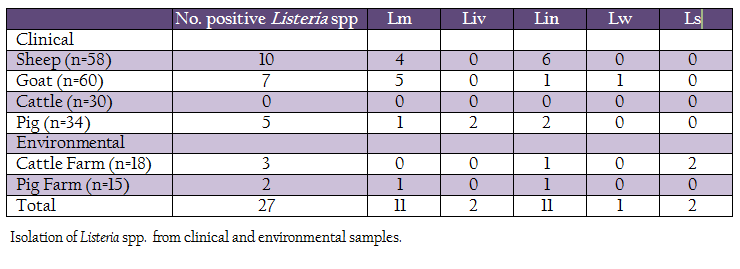
A total of 215 samples were collected from Konkan region of Maharashtra, India (Table 1). Of these, 182 samples (vaginal swabs, blood, milk and faeces) were collected from domestic animals such as sheep, goat, bovines and pig, while, 33 samples were collected from cattle and pig farms surroundings (soil and floor swabs). The samples were collected in sterile containers, transported to laboratory on ice and processed for isolation of listerial spp.
Isolation of Listeria Species
Isolation of Listeria spp. was attempted as per USDA–FSIS method (USDA, 2013). The samples (blood, milk, faeces and soil) were inoculated (approx. 5 ml/5 g) into 45 ml of University of Vermont Medium (UVM)–1 supplemented with acriflavin and nalidixic acid, and incubated at 37oC for 18–24 h. Vaginal and floor swabs were directly inoculated in 10 ml UVM broth. Further enrichment of the samples was carried by inoculating 0.1 ml of UVM–1 to 10 ml of UVM–2 broth. Inoculated UVM–2 broth was incubated further for 24 h at 370C. A loopful of enriched UVM–2 broth was streaked directly on PALCAM agar for selective isolation of listerial colonies. The inoculated agar plates were incubated at 37oC for 48 h. The isolated pinpoint grayish–green colonies surrounded by black zone of esculin hydrolysis were presumed as Listeria. These colonies were further purified on PALCAM agar and stored in refrigerated conditions in BHI broth.
Biochemical Characterization and Identification of Isolates
A single isolated colony from PALCAM agar was inoculated in fresh BHI broth and incubated at 37oC for 18 h. The freshly grown culture was then studied for their morphological and biochemical characters. Morphology was observed under light microscope while, Listeria specific biochemical tests such as catalase, oxidase, characteristics tumbling motility at 20–25 C and fermentation of sugars (rhamnose, xylose, mannitol and α–methyl D – mannopyranoside) were performed. The isolates were compared with standard Listeria spp. for identification. Isolates suspected as L. monocytogenes from biochemical tests were further subjected to specific tests such as hemolysis on sheep blood agar (SBA), Christie, Atkins, Munch–Petersen (CAMP) test and phosphatidylinositol phospholipase C activity (PI–PLC) (Gorski et al., 2008).
Genotypic Characterization
Biochemically confirmed L. monocytogenes isolates were subjected to serotyping by multiplex PCR method as described by Doumith et al. (2004). The genomic DNA was extracted using DNA isolation kit (Chromos Biotech, India). Oligonucleotide primers (Doumith et al., 2004) were synthesized from Sigma. The 50 µl PCR mixture contained 2.5μl of 10x PCR buffer (100 mM Tris–HCl, pH 8.3 at 25°C; 500 mM KCl; 15 mM MgCl2; 0.01% gelatin), 2 mM dNTP mix, 3 mM MgCl2 and 0.3 μM each of forward and reverse primers (lmo0737, lmo2819, lmo2110 and prs gene), 2 units of Taq DNA polymerase and 50 ng of DNA template. The final volume was adjusted by sterilized deionised water. The reaction was performed in a Mastercycler epGradient (Eppendorf, Germany) with a preheated lid. PCR was performed with an initial denaturation step at 94°C for 3 min; 35 cycles of 94°C for 0.40 min, 53°C for 1.15 min, and 72°C for 1.15 min; and one final cycle of 72°C for 7 min. Five microliters of the reaction mixture was mixed with 1μl of gel loading buffer and separated on a 1.5% agarose gel pre–stained with ethidium bromide. The gel was visualized under Alpha–Imager gel doc system.
PCR amplification of the hlyA gene was performed as described by (Paziak–Domańska et al., 1999). The 50 µl PCR mixture contained 2.5μl of 10x PCR buffer (100 mM Tris–HCl, pH 8.3 at 25°C; 500 mM KCl; 15 mM MgCl2; 0.01% gelatin), 2 mM dNTP mix, 2 mM MgCl2 and 0.3 μM of forward and reverse primers, 1.5 units of Taq DNA Polymerase and 50ng of DNA template. The final volume was adjusted by sterilized deionised water. The reaction mixture was subjected to an initial denaturation at 950C for 2 min followed by 35 cycles each of 15 s denaturation at 950C, 30 s annealing at 600C and 1 min 30 s extensions at 720C. It was followed by final extension of 10 min at 720C and held at 40C. The PCR products were tested as described above.
Antibiotic Sensitivity of Isolates
Disk diffusion susceptibility tests were performed according to the standard reference procedure of the Clinical and Laboratory Standards Institute (Anon, 2006; Altuntas et al., 2012). A single well–isolated colony of L. monocytogenes was transferred into 10 ml BHI broth, incubated at 37°C for 24 h, diluted 1:10 in 9 ml 0.1% peptone water to a turbidity equivalent to 0.5 McFarland standard, and spread on surface of Mueller–Hinton Agar (MHA) plate. Antibiotic discs of vancomycin (VA10), ciprofloxacin (Cf5), erythromycin (E15), gentamycin (G10), penicillin–G (P10), sulphazidine (Sz 30), amphicilin (A10), oxytetracycline (O30), chloramphenicol (C30) and doxycycline (D30) were placed on the surface of each inoculated MHA plate. After incubation for 24 h at 37°C, the diameter (in mm) of the zone around each disk was measured and interpreted in accordance with the Clinical and Laboratory Standards Institute Standards guidelines (Anon et al., 2006) to classify the antibiotic sensitivity of each isolate. Staphylococcus aureus ATCC 6538 was used as standard strain.
RESULTS
Isolation and Identification of Listeria
Out of 215 samples (clinical and farm environment) collected, 27(12.55%) samples were found positive for Listeria spp. After further characterization, 11 (5.11%) isolates were confirmed as L. monocytogenes, 2 (0.93%), L. ivanovii, 11 (5.11%), L. innocua, 1 (0.46%), L. welshimeri and 2 (0.93%) as L. seeligeri (Table 1). Except clinical samples collected from cattle, all other types of samples showed presence of Listeria spp. Four L monocytogenes were isolated from clinical samples collected from sheep, five from goats, one each from pig and pig–associated environment. The L. ivanovii was isolated from a clinical case in pig. No L. monocytogenes or L. ivanovii could be isolated from cattle and associated environment.
Biochemically identified L. monocytogenes and L. ivanovii spp. were further characterized by phenotypic assays. All the L. monocytogenes and L. ivanovii exhibited weak hemolysis. In CAMP test, L. monocytogenes and L. ivanovii showed increased zone of hemolysis toward Staphylococcus aureus (MTCC 1144) and Rhodococcus equi (MTCC 1135), respectively. All the Listeria spp. were tested on ALOA medium to determine the ß–glucosidase and ability to produce the phospholipase enzymes. All the isolates showed blue–green color zone confirming the Listeria spp. while in addition, L. monocytogenes and L. ivanovii exhibited an opaque white halo of hydrolysis of phosphotidyliositol or lethicin in the medium due to phospholipases. Presence of phospholipase activity was considered as indicator the pathogenicity.

Figure 1: Multiplex PCR serotyping for determination of the serogroups of the isolates; Lane 1: Isolate FC2 L; monocytogenes serogroup 4b, 4d, 4c; Lane 2: 16 L. monocytogenes serogroup: 4b, 4d, 4c; Lane 3: 13 L. monocytogenes serogroup 4b, 4d, 4c; Lane 4: 22 L. monocytogenes serogroup 1/2b, 3b, 7; Lane 5:19 L. monocytogenes serogroup 1/2b, 3b, 7; Lane 6: Listeria spp., Positive control (PC); L. monocytogenes MTCC 1143; NC: Negative control; M: 100 bp DNA ladder.
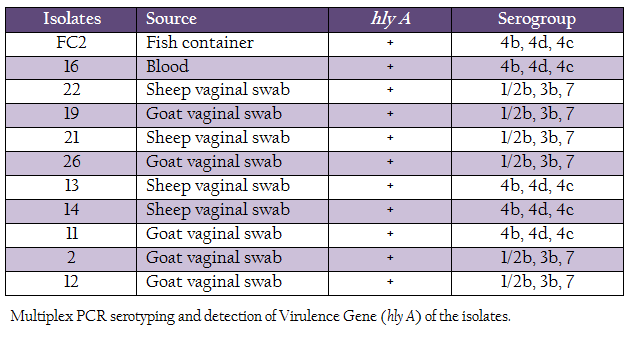
L. monocytogenes isolates were characterized for the presence of the hemolysin gene and by multiplex PCR based–serotyping. Multiplex PCR based serotyping revealed 5 isolates as serotype 4b while remaining 6 were of serotype 1/2b (Table 2, Figure 1). All the L. monocytogenes isolates were found to possess hlyA gene, which is responsible for hemolysin. In case of L. ivanovii genus, specific prs gene was amplified.
Antibiotic Sensitivity Testing
L. monocytogenes isolates were checked for their susceptibility towards the commonly prescribed antibiotics. All isolates were sensitive towards, erythromycin, oxytetracycline, ampicillin, doxycycline and ciprofloxacin and showed intermediate resistances towards the chloramphenicol, penicillin, gentamicin and vancomycin. However, a single L. monocytogenes serotype 4b isolate from pig placental tissue exhibited multi–antibiotic resistance. All 10 L. monocytogenes isolates shown resistance to sulphazidine.
DISCUSSION
Listeria spp. are ubiquitous in nature and has been isolated from wide environmental sources (Liu, 2008). The organism possesses ability to survive in harsh conditions and therefore can persist in environment. Because of such persistence Listeria spp. can easily enter in food chain (Carpentier and Cerf, 2011). Of the known Listeria spp., L. monocytogenes is pathogenic to humans and animals, while L. ivanovii is mainly an animal pathogen. Several foodborne outbreaks are attributed to L. monocytogenes (Ramaswamy et al., 2007). Therefore, food quality controlling authorities from several developed countries have enforced strict regulations over occurrence of L. monocytogenes in food and food products (CDC, 2012). However, such regulations are largely lacking in developing countries because of underestimated listerial scenario. In India, due to lack of awareness, burden of other traditional diseases, rare expertise, and poor reporting, the incidence of listeriosis is unknown (Dandona et al., 2004; Dandona et al., 2009). To understand the listeriosis in detail, there is a need of systematic and coordinated studies to estimate the prevalence of L. monocytogenes in different habitats, occurrence of listeriosis in humans as well as in animals and their concordance of occurrence and actual disease. The epidemiological studies would help in understanding of the sources of infection and persistence and their risk assessment, routes of transmission, clinical forms and allow for better management of the listerial infection. Present study was performed to determine the prevalence of Listeria spp. from Konkan region of Maharashtra, India. The Konkan region contributes a great source of animal and crop originated food. Listeria spp. were isolated from 12.55% samples. L. monocytogenes and L. ivanovii were isolated from 5.11% and 0.93% samples, respectively. The L. monocytogenes isolates were mainly isolated from sheep and goats. A bacterial etiology studied in relation to abortions by over 203 sheep and goat flocks showed 5–6% of L. monocyotgenes infections (Sharma et al., 2008). Barbuddhe et al. (2000) have reported presence of L. monocytogenes in 6.66% and 7.4% meat samples of goats. In India, L. monocytogenes has been reported from fish (Dhanashree et al., 2003; Gawade et al., 2011; Parihar et al., 2008; Das et al., 2012), meat (Doijad et al., 2010; Bramhabhatt and Anjaria, 1993), and milk (Doijad et al., 2011; D’Costa et al., 2012). In the present study, L. monocytogenes could not be isolated from cattle. L. ivanovii was isolated from pig samples. Presence of L. monocytogenes in animal associate–environment may lead to cause infection to animals being reared in that area, however, it warrants future investigations.
There are 12 serotypes of L. monocytogenes known till date, of which serotypes 1/2a, 1/2b and 4b contribute more than 98% of outbreaks (Cheng et al., 2008). Our serotyping data confirmed 6 isolated to be serotype 1/2b and 5 as serotype 4b. L. monocytogenes serotype 4b has been considered as cosmopolitan (Herd and Kocks, 2001; Cheng et al., 2008). Presence of such outbreak–associated serotype is of public health concern. However, more studies with large number from humans and animal clinical cases may present the actual scenario.
The antibiotic resistance of the pathogen is a significant public health concern. Recent reports suggest the evolution of L. monocytogenes towards antibiotic resistance (Charpentier and Courvalin 1999; Altuntas et al., 2012; Soni et al., 2013). It is suggested that the increased use of antibiotics for therapeutic purposes in animals and humans may lead to the development of antibiotic resistance (Palumbo et al., 2010; Yan et al., 2010). Depending upon different geographical area, antibiotic resistance patterns of L. monocytogenes in food and environmental sources may change (Yan et al., 2010). We tested all L. monocytogenes isolates for their antibiotic sensitivity. A single L. monocytogenes serotype (4b) isolates from pig placental tissue exhibited multi–antibiotic resistance while remaining 10 isolates were sensitive towards the majority of the antibiotics studied. Interestingly, all the L. monocytogenes isolates were resistant to sulphadiazine. The study conducted by Dhanashree et al., (2003) found similar results wherein L. monocytogenes isolates which were sensitive towards commonly used antibiotics were reported.
The present study showed the prevalence of L. monocytogenes in clinical and environmental samples from Konkan region of Maharashtra, India. The isolates belonged to serotypes having epidemic potential. There is a possibility of transfer of this pathogen from animals to foods of animal origin and thereafter the food chain.
ACKNOWLEDGEMENTS
We thank Director, ICAR Research Complex for Goa, Old Goa for providing laboratory facilities. The work was supported by grants from Department of Biotechnology, Government of India.
REFERENCES
Adhikary R, Joshi S (2010). Neonatal listeriosis: a case report from sub–Himalayas. Indian. J. Med. Microbiol. 29(1): 79.
http://dx.doi.org/10.4103/0255-0857.76536
PMid:21304207
Altuntas EG (2012). Antibiotic and bacteriocin sensitivity of Listeria monocytogenes strains isolated from different foods. Food Nutri.Sci. 3(3): 363 – 368.
http://dx.doi.org/10.4236/fns.2012.33052
Barbuddhe S, Malik SVS, Bhilegaonkar KN, Kumar P, Gupat LK (2000). Isolation of Listeria monocytogenes and anti–listeriolysin O detection in sheep and goats. Small. Rumin. Res. 38(2): 151 – 155.
http://dx.doi.org/10.1016/S0921-4488(00)00155-3
Barbuddhe SB, Hain T, Chakraborty T (2008). The Genus Listeria. In:Practical Handbook of Microbiology, CRC Press, Boca Raton. 533 –562.
http://dx.doi.org/10.1201/9781420009330.ch34
Bertsch D, Rau J, Eugster MR, Haug MC, Lawson PA, Lacroix C, Meile L (2013) Listeria fleischmannii sp. nov., isolated from cheese. Int. J. Syst. Evol. Microbiol. 63(2): 526 – 532.
http://dx.doi.org/10.1099/ijs.0.036947-0
PMid:22523164
Bramhabhatt MN, Anjaria JM (1993). Analysis of market meat for possible contamination with Listeria monocyotgenes. Indian J. Anim. Sci. 63: 687.
Carpentier B, Cerf O (2011). Review–Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food. Microbiol. 145(1): 1 – 8.
http://dx.doi.org/10.1016/j.ijfoodmicro.2011.01.005
PMid:21276634
CDC (2012). Multistate Outbreak of listeriosis linked to whole cantaloupes from Jensen Farms, Colorado. Available at: http://www.cdc.gov/listeria/outbreaks [Accessed December 20, 2013].
Charpentier E, Courvalin P (1999). Antibiotic resistance in Listeria spp. Antimicrob. Agents. Chemother. 43(9): 2103 – 2108.
PMid:10471548 PMCid:PMC89430
Charpentier E, Courvalin P (1999). Antibiotic Resistance in Listeria spp . Antimicrob. Agents. Chemother. 43(9): 2103 – 2108.
PMid:10471548 PMCid:PMC89430
Cheng Y, Robin S, Kathariou S (2008). Genomic Division/Lineages, Epidemic clones, and Population Structure. In: Hnadbook of Listeria monocytogenes, CRC press, Boca Raton. 337 – 358.
D'Costa D, Bhosale SN, Dhuri RB, Kalekar S, Rodrigues J, Doijad S, Barbuddhe SB (2012). Occurrence and characterization of Listeria species isolated from the milk production chain. Milchwissenschaft. 67(1): 43 – 46.
Dandona L, Sivan YS, Jyothi MN, Bhaskar VS, Dandona R (2004). The lack of public health research output from India. BMC Public Health. 4:55.
http://dx.doi.org/10.1186/1471-2458-4-55
PMid:15563377 PMCid:PMC539252
Dandona L, Raban MZ, Guggilla RK, Bhatnagar A, Dandona R. (2009). Trends of public health research output from India during 2001 – 2008. BMC Med. 7:59.
http://dx.doi.org/10.1186/1741-7015-7-59
PMid:19828017 PMCid:PMC2766381
Das S, Lalitha KV, Thampuran N, Surendra PK (2012). Isolation and characterization of Listeria monocytogenes from tropical seafood of Kerala, India. Ann. Microbiol. 63(3): 1093 – 1098
http://dx.doi.org/10.1007/s13213-012-0566-9
Dhama K, Verma AK, Rajagunalan S, Kumar A, Tiwari R, Chakraborty S, Kumar R (2013). Listeria monocytogenes infection in poultry and its public health importance with special reference to food borne zoonoses, Pak. J. Biol. Sci. 16(7): 301 – 308.
http://dx.doi.org/10.3923/pjbs.2013.301.308
PMid:24498796
Dhanashree B, Otta SK, Karunasagar I, Goebel W, Karunasagar I (2003). Incidence of Listeria spp. in clinical and food samples in Mangalore, India. Food. Micobiol. 20(4): 447 – 453.
http://dx.doi.org/10.1016/S0740-0020(02)00140-5
Deb R, Kumar A, Chakraborty S, Verma AK, Tiwari R, Dhama K, Singh U, Kumar S (2013). Trends in diagnosis and control of bovine mastitis: A review. Pak. J. Biol. Sci. 16(23): 1653 – 1661.
http://dx.doi.org/10.3923/pjbs.2013.1653.1661
PMid:24506032
Doijad S, Barbuddhe SB, Garg S, Kalekar S, Rodrigues J, Decosta D, Bhosale S, Chakraborty T (2011). Incidence and genetic variability of Listeria species from three milk processing plants. Food Cont. 22(12): 1900 –1904.
http://dx.doi.org/10.1016/j.foodcont.2011.05.001
Doijad S, Vaidya V, Garg S, Kalekar S, Rodrigues J, Decosta D, Bhosale S, Barbuddhe SB (2010) Isolation and characterisation of Listeria spp. from raw and processed meats. J.Vet. Public Hlth. 8(2): 83 – 88.
Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P (2004). Differentiation of the Major Listeria monocytogenes Serovars by Multiplex PCR. J. Clin. Microbiol. 42(8): 3819 –3822.
http://dx.doi.org/10.1128/JCM.42.8.3819-3822.2004
PMid:15297538 PMCid:PMC497638
Farber JM, Peterkin PI (1991). Listeria monocytogenes, a food–borne pathogen. Microbiol. Rev. 55(3): 476 – 511.
PMid:1943998 PMCid:PMC372831
Gawade L, Barbuddhe SB, Bhosle S (2011). Isolation and Confirmation of Listeria Species from Seafood off Goa Region by Polymerase Chain Reaction. Indian J.Microbiol. 50(4): 385 – 389.
http://dx.doi.org/10.1007/s12088-011-0064-y
PMid:22282604 PMCid:PMC3209847
Goulet V, Hebert M, Hedberg C, Laurent E, Vaillant V, De Valk H, Desenclos JC (2012). Incidence of listeriosis and related mortality among groups at risk of acquiring listeriosis. J. Microbiol. Methods. 54(5): 652 – 660.
Graves LM, Helsel LO, Steigerwalt AG, Morey RE, Daneshvar MI, Roof SE, Orsi RH, Fortes ED, Milillo SR, den Bakker HC, Wiedmann M, Swaminathan B, Sauders BD. (2010). Listeria marthii sp . nov ., isolated from the natural environment, Finger Lakes National Forest. Int. J. Syst. Evol. Microbiol. 60 (6): 1280 – 1288.
http://dx.doi.org/10.1099/ijs.0.014118-0
PMid:19667380
Guillet C, Lambert JO, Le Monnier A, Leclercq A, Mechaï F, Bruneel MMF, Bielecka MK, Scortti M, Disson O, Berche P, Boland VJ, Lortholary O, Lecuit M. (2010). Human Listeriosis Caused by Listeria ivanovii. Emerg. Infect. Dis. 16(1): 136 – 138.
http://dx.doi.org/10.3201/eid1601.091155
PMid:20031061 PMCid:PMC2874378
Halter EL, Neuhaus K, Scherer S (2013). Listeria weihenstephanensis sp . nov ., isolated from the water plant Lemna trisulca taken from a freshwater pond. Int. J. Syst. Evol. Microbiol. 63(2): 641 – 647.
http://dx.doi.org/10.1099/ijs.0.036830-0
PMid:22544790
Herd M, Kocks C (2001). Gene fragments distinguishing an epidemic–associated strain from a virulent prototype strain of Listeria monocytogenes belong to a distinct functional subset of genes and partially cross–hybridize with other Listeria species. Infect. Immun. 69(6): 3972 –3979.
http://dx.doi.org/10.1128/IAI.69.6.3972-3979.2001
PMid:11349066 PMCid:PMC98459
Jallewar PK, Kalorey DR, Kurkure NV, Pande VV, Barbuddhe SB (2007). Genotypic characterization of Listeria spp. isolated from fresh water fish. Int. J. Food Microbiol. 114(1): 120 – 123.
http://dx.doi.org/10.1016/j.ijfoodmicro.2006.09.034
PMid:17182144
Kalorey DR, Kurkure NV, Warke SR, Rawool DB, Barbuddhe SB (2008). Listeria species in bovine raw milk: A large survey of Central India. Food Cont. 19(2): 109 – 112.
http://dx.doi.org/10.1016/j.foodcont.2007.02.006
Kathariou S (2002). Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J Food Prot. 65(11): 1811 – 1829.
PMid:12430709
Kvistholm J (2010). Substantial increase in listeriosis, Denmark 2009. Euro. Surveill. 15(12):1 – 4.
Leclercq A, Clermont D, Bizet C, Grimont PA, Le Flèche–Matéos A, Roche SM, Buchrieser C, Cadet–Daniel V, Le Monnier A, Lecuit M, Allerberger F. (2010). Listeria rocourtiae sp. nov. Int. J. Syst. Evol. Microbiol. 60(9): 2210 – 2214.
http://dx.doi.org/10.1099/ijs.0.017376-0
PMid:19915117
Liu D (2008). Epidemiology:In Handbook of Listeria monocytogenes. CRC press, Boca Raton : 27 – 60.
http://dx.doi.org/10.1093/aje/kwn280
http://dx.doi.org/10.1097/EDE.0b013e3181632c24
http://dx.doi.org/10.1093/aje/kwn398
http://dx.doi.org/10.1016/j.annepidem.2008.05.005
http://dx.doi.org/10.1093/aje/kwn227
http://dx.doi.org/10.1097/EDE.0b013e3181690731
http://dx.doi.org/10.1093/aje/kwn396
http://dx.doi.org/10.1093/aje/kwn339
Low JC, Donachie W (1997). A review of Listeria monocytogenes and listeriosis.Vet J. 153(1): 9 –29.
http://dx.doi.org/10.1016/S1090-0233(97)80005-6
Lyytikäinen O, Nakari UM, Lukinmaa S, Kela E, Nguyen TMN, Siitonen A (2006). Surveillance of listeriosis in Finland during 1995–2004. Euro. Surveill. 11(6): 82 – 85.
PMid:16801696
Manoj YB, Rosaling GM, Karunasagar I (1991). Listeria spp. in fish and fish–handling areas, Mangalore, India. Asian Fish. Sci. 4: 119 – 122.
Moharem AS, Raj APC, Janardhana GR (2007). Incidnes of Listeria species in seafood of Mysore, India. J. Food Safy. 27(4): 362 – 372.
http://dx.doi.org/10.1111/j.1745-4565.2007.00085.x
Mokta KK, Kanga AK, Kaushal RK (2010). Neonatal listeriosis: a case report from sub–Himalayas. Indian. J. Med. Microbiol. 28(4): 385 – 387.
http://dx.doi.org/10.4103/0255-0857.71824
PMid:20966576
Palumbo D, Iannaccone M, Porta A, Capparelli R (2010). Experimental antibacterial therapy with puroindolines, lactoferrin and lysozyme in Listeria monocytogenes–infected mice. Microbes. Infect. 12(7): 538 –545.
http://dx.doi.org/10.1016/j.micinf.2010.03.010
PMid:20348006
Parihar VS, Barbuddhe SB, Danielsson TML, Tham W (2008). Isolation and characterization of Listeria species from tropical seafoods. Food Control 19(6): 566 – 569.
http://dx.doi.org/10.1016/j.foodcont.2007.06.009
Paziak DB, Bogusławska E, Wieckowska SM, Kotłowski R, Rózalska B, Chmiela M, Kur J, Dabrowski W, Rudnicka W. (1999). Evaluation of the API test, phosphatidylinositol–specific phospholipase C activity and PCR method in identification of Listeria monocytogenes in meat foods. FEMS Microbiol. Lett. 171(2): 209 – 214.
http://dx.doi.org/10.1111/j.1574-6968.1999.tb13434.x
Ramaswamy V, Cresence VM, Rejitha JS, Lekshmi MU, Dharsana KS, Prasad SP, Vijila HM. (2007). Listeria–review of epidemiology and pathogenesis. J. Microbiol. Immunol. Infect. 40(1): 4 – 13.
PMid:17332901
Rocourt J, Jacquet C, Reilly A (2000). Epidemiology of human listeriosis and seafoods. Int. J. Food Microbiol. 62(3): 197 – 209.
http://dx.doi.org/10.1016/S0168-1605(00)00336-6
Sharma M, Batta MK, Katoch RC, Andersen AA (2008). A field investigation of bacterial etiology of abortions among migratory sheep and goats in North–West hill states of India. Vet. Arhiv. 78(1): 65 – 71.
Soni DK, Singh RK, Singh DV, Dubey SK (2013). Characterization of Listeria monocytogenes isolated from Ganges water, human clinical and milk samples at Varanasi, India. Infect. Genet. Evol. 14: 83 – 91.
http://dx.doi.org/10.1016/j.meegid.2012.09.019
PMid:23201044
USDA. (2013). Laboratory Guidebook : Isolation and Identification of Listeria monocytogenes from Red Meat, Poultry and Egg Products, and Environmental Samples. USDA.
Yadav MM, Roy A, Bhanderi B, Jani RG (2011). Prevalence of Listeria species including L . monocytogenes from apparently healthy animals at Baroda Zoo, Gujarat State, India. J. Threat Taxa 3(7): 1929 – 1935.
http://dx.doi.org/10.11609/JoTT.o2094.1929-35
Yan H, Neogi SB, Mo Z, Guan W, Shen Z, Zhang S, Li L, Yamasaki S, Shi L, Zhong N (2010). Prevalence and characterization of antimicrobial resistance of foodborne Listeria monocytogenes isolates in Hebei province of Northern China, 2005–2007. Int. J. Food Microbiol. 144(2): 310 – 316.
http://dx.doi.org/10.1016/j.ijfoodmicro.2010.10.015
PMid:21074885