Advances in Animal and Veterinary Sciences
Research Article
Cross Sectional Study for Evaluation of Rapid Test and RT-PCR in Detection of BRV in Fecal Samples of Diarrhetic Calves in Egypt
Ebtsam A. Abouelyazeed, Rabab T. Hassanien, Ahmed F. Afify*
Virology Research Department, Animal Health Research Institute, Agriculture Research Center (ARC), Dokki, Giza, Egypt.
Abstract | In the present study, forty-three fecal samples were collected between 2018 and 2020, were tested for bovine Rotavirus (RBV) by rapid commercial strip test, Ag detection sandwich ELISA and RT-PCR for screening and matching of sensitivity and specificity. Our results revealed that seven samples (7/43; 16.2%) were positive by rapid test, fourteen and ten samples were positive by Ag detection ELISA (14/43; 32.5%) and RT-PCR (10/43; 23.2%), respectively. Sequence analysis for one positive sample showed its complete identity with the recent BRV Egyptian strains which reassures the vaccine reliability and negates any recent virus evolution. Meanwhile, comparison between both the rapid test and RT-PCR and ELISA, the sensitivity was 50% and 71.5% respectively, while specificity was identical. In conclusions, ELISA can be considered as a simple, sensitive and reliable test for BRV detection and devoid the drawbacks of other tests.
Keywords | Bovine rotavirus, Rapid test, Gastroenteritis
Received | October 10, 2020; Accepted | December 03, 2020; Published | January 15, 2021
*Correspondence | Ahmed Fawzy Afify, Virology Research Department, Animal Health Research Institute, Agriculture Research Center (ARC), Dokki, Giza, Egypt; Email: Fieldagy_ahmed@yahoo.com
Citation | Abouelyazeed EA, Hassanien RT, Afify AFM (2021). Cross sectional study for evaluation of rapid test and RT-PCR in detection of BRV in fecal samples of diarrhetic calves in Egypt. Adv. Anim. Vet. Sci. 9(3): 387-392.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.3.387.392
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Abouelyazeed et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Neonatal calf diarrhea is one of the most common animal health concerns for dairy industry, which has been responsible for high morbidity and mortality in calf (Pardo-Mora et al., 2018). In cattle, rotavirus is one of the main pathogens involved in the development of Bovine Neonatal Diarrhea (BND). Group-A rotaviruses are responsible of calves’ diarrhea, which lead to 5-20% calves losses (Kim et al., 2011). The name rotavirus stems from the characteristic wheel-like appearance of the virus when observed by electron microscopy, which is derived from the Latin word “rota”meaning “wheel”. Rotavirus belongs to family Reoviridae, subfamily Sedoreovirinae genus Rotaviruses, non-enveloped virus with a diameter of 65-70 nm, characterized by segmented genomes comprising of 11 segments of double stranded RNA which is enclosed in triple layered icosahedral capsid layer, its genome encodes six SP (structural proteins) (VP1, VP2, VP3, VP4, VP6, and VP7) and six NSP (NSP1 to NSP6) (Desselberger, 2014; Zhou et al., 2015). Rotaviruses are species-specific, but cross-species transmission might occur due to its in vitro implementation. Likewise, comparison of genetic data between human and animal rotaviruses revealed their close identity (Cook et al., 2004).
Different diagnostic approaches are used to detect the rotavirus that include electron microscope (Saif et al., 1980; Reynolds et al., 1984), virus isolation (Gulyaz et al., 2010), immunochromatographic rapid diagnostic assays (Al-Yousif et al., 2001; Klein et al., 2009; Altug et al., 2013), Enzyme-linked immunosorbent assay (ELISA) (Phillips et al., 2009; Gulyaz et al., 2010) and RT-PCR (Aich et al., 2007; Asano et al., 2010).
The aim of this study is to screen the prevalence of BRV among some dairy herds with enteric disease in Egypt during 2018-2020. After that, sensitivity and specificity comparison between the rapid test and RT-PCR with ELISA as a standard test for detection of BRV in fecal samples.
MATERIALS AND METHODS
Ethical approval
International, national, and/or institutional guidelines for the samples collection were followed with the approval of the Local Ethics Committee on Animal Experimentation in Animal Health Research Institute, Agriculture Research Center (ARC), Egypt.
Samples collection and preparation
A total of forty- three fecal samples were collected from calves accomplished with acute gastroenteritis aged up to three months in Egypt between 2018 and 2020. Samples were transferred to Virology department, Animal health research institute, for laboratory diagnosis.
Fecal samples were prepared either a 10% (W/V) suspension of solid or semisolid feces in 0.01 M phosphate buffered saline (PBS) (pH 7) or as a 20% (V/V) suspension of liquid feces in 0.01 M PBS (pH 7). All samples were centrifuged at 1500 xg/ 5 minutes, and the supernatants were collected for testing then stored at -80°C for further usage (Barry et al., 2009).
Detection of BRV antigen using rapid test and ELISA assay
Fecal samples were tested for BRV antigen using commercial strip test kits (BIO K 384 – Dip-Fit bovine Rotavirus®, Bio-X Diagnostics, SA., Belgium) following the manufacturer’s instructions. Likewise, commercial sandwich ELISA kit (Monoscreen Ag Elisa®, Bio-X Diagnostics, SA., Belgium) following manufacturer’s instructions. The optical density (OD) for each well was measured at 450 nm. Results were interpreted according to manufacturer’s instructions (Beksisa et al., 2020).
RNA extraction and Reverse transcriptase polymerase chain reaction (RT-PCR)
RNA extraction was performed using the QIAamp viral RNA Mini kit (Qiagen®, GmbH, Germany) according to the manufacturer’s instructions. Specific primers targeted the NSP5 gene of Bovine Rotavirus (Sense primer: GAT ATT GGA CCA TCT GAT TCT GCT TCA AA, antisense primer: GAA ATC CAC TTG ATC GCA CCC AA) (Schroeder et al., 2012) supplied by Metabion©, Germany. The RT-PCR reaction was performed in a 25 µl reaction containing 12.5 µl of Quantitect probe rt-PCR buffer (QIAgen®, Gmbh, Germany), 1 µl of sense primer (20 pmol concentration), 1 µl of anti-sense primer (20 pmol concentration), 0.25 µl of rt-enzyme, 4.25 µl of nuclease free water, and 6 µl of RNA template. The cycling reaction was performed in a Biometra thermal cycler (Biometra TRIO 48©, Analytik Jena, Germany). Reverse transcription was carried out at 50°C for 30 min, then initial denaturation step at 95°C for 5 min, followed by 35 cycles of 94°C for 30 sec., 60°C for 45 sec., 72°C for 45 sec. and final extension step at 72°C for 10 min. (Schroeder et al., 2012).
Sequencing, sequence analysis and phylogeny
The PCR products were separated by electrophoresis on 1.5% agarose (Applichem®, GmbH, Germany). The gel was photographed by a gel documentation system (Alpha Innotech, Biometra©) and the data was analyzed through computer software. The PCR products were purified using QIAquick PCR Product purification kit (Qiagen, Valencia, Germany). Bigdye Terminator V3.1 cycle sequencing kit (Perkin-Elmer) was used for sequencing using Applied Biosystems genetic analyzer (HITACHI, Japan), a BLAST® analysis (Basic Local Alignment Search Tool) was initially performed to check the sequence identity (Altschul et al., 1990). The phylogenetic tree was constructed by the MegAlign module of Lasergene DNA Star version 12.1 using maximum likelihood (Tamura et al., 2013).
RESULTS AND DISCUSSION
Bovine rotavirus is a major causative agent for acute gastroenteritis among calves. So, availability of simple, rapid, specific, and sensitive diagnostic approach for detection of viral agents associated with enteric disease is an urgent necessity and will help in control and eradication of the disease in newborn calves (Theil, 1990). ELISA assay is widely used in diagnostic facilities for Rotavirus antigen because it provides rapid detection compared to other tests. Meanwhile, it is a WHO gold standard test in developing countries, very sensitive for gp A of rotavirus detection in several mammalian species (Phillips et al., 2009).
In the present study, we used three comparative tests (ELISA, rapid strip test and RT-PCR) for detection of BRV antigen in forty-three fecal samples collected from diseased calves showing suspect signs to BRV.
Bovine rotavirus was detected in seven samples by Rapid test (16.2%), in fourteen samples by Ag detection ELISA (32.5%) and in ten samples by RT-PCR (23.2%) (Figure 1), comparison between the results of the three tests is shown in Table 1.
Five samples (11.6%) were positive by the three assays, whilst twenty nine samples were negative using the same assays (67.4%). Both Rapid test and ELISA succeeded to detect two samples (4.6%) failed to be identified by RT-PCR. On the other hand, both ELISA and RT-PCR detected five samples (11.6%) which gave negative results with the Rapid test. In addition, two samples (4.6%) were positive by ELISA only.
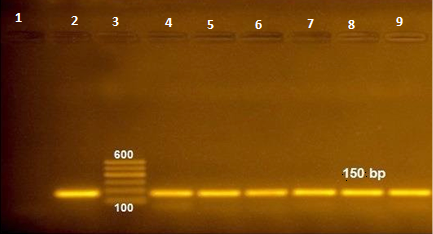
Figure 1: RT-PCR amplified products of NSP5 gene at 150 bp (lane 1: negative control, lane 2: positive control, lane 3: 100 bp ladder and lane 4-9 positive samples).
Table 1: Comparison of outcomes from three testing methods for detection of BRV in 43 fecal samples a.
|
Result of test b |
Frequency (No. of specimen) |
Πi | ||
| ELISA | Rapid test | RT-PCR | ||
| + | + | + | 5 | 0.116 |
| - | - | - | 29 | 0.674 |
| + | + | - | 2 | 0.046 |
| + | - | + | 5 | 0.116 |
| + | - | - | 2 | 0.046 |
a: Agreement was determined with reference to the total number of specimens tested (n = 43). Both the Rapid test and ELISA were used for detecting BRV in calf feces. πi =frequency/n; b: + means positive and - means negative.
Sensitivity and specificity of both RT-PCR and Rapid test were calculated against ELISA standard test. Both tests were 100% specific; however, Rapid test was 50% sensitive and RT-PCR had 71.4% sensitivity when compared to ELISA (as shown in Tables 2 and 3).
P value was calculated for both Rapid test and RT-PCR using Chi square values, Value of P<0.001 which means that there are significant differences between the results of both Rapid test and RT-PCR in comparison to standard test (ELISA).Our findings suggest using BRV rapid test as a field test only as it has same specificity with standard test (100%), so true positive results is guaranteed, but cannot be adopted for laboratory diagnosis due to low sensitivity, which may be misleading because of 50% false negative suspected results because of the sampling is carried out in the late course of the disease when the level of virus shedding is decreased and the amount of viral particles is low (Uyunmaz et al., 2019). Likewise, our results are similar to previous findings that revealed that the rotavirus rapid immunochromatographic test had greater specificity (100%), but lower sensitivity (57%) in comparison with ELISA test in feces samples from 60 calves Luginbühl et al. (2005).
Table 2: Sensitivity and specificity of Rapid test compared with ELISA as reference methods for detection of BRV infection a.
| Rapid test | Total | ||||
| Positive | Negative | ||||
| ELISA | Positive | Count | 7 | 7 | 14 |
| % within ELISA |
50.0%b |
50.0% | 100.0% | ||
| % within Rapid test | 100.0% | 19.4% | 32.6% | ||
| Negative | Count | 0 | 29 | 29 | |
| % within ELISA | 0.0% |
100.0%c |
100.0% | ||
| % within Rapid test | 0.0% | 80.6% | 67.4% | ||
| Total | Count | 7 | 36 | 43 | |
| % within ELISA | 16.3% | 83.7% | 100.0% | ||
| % within Rapid test | 100.0% | 100.0% | 100.0% | ||
a: n=43; b: Sensitivity=[TP/(TP+FN)]*100; c: Specificity=[TN/(TN+FP)]*100.
Table 3: Sensitivity and specificity of RT-PCR compared with ELISA as reference methods for detection of BRV infection a
| RT-PCR | Total | ||||
| Positive | Negative | ||||
| ELISA | Positive | Count | 10 | 4 | 14 |
| % within ELISA |
71.4%b |
28.6% | 100.0% | ||
| % within RT-PCR | 100.0% | 12.1% | 32.6% | ||
| Negative | Count | 0 | 29 | 29 | |
| % within ELISA | 0.0% |
100.0%c |
100.0% | ||
| % within RT-PCR | 0.0% | 87.9% | 67.4% | ||
| Total | Count | 10 | 33 | 43 | |
| % within ELISA | 23.3% | 76.7% | 100.0% | ||
| % within RT-PCR | 100.0% | 100.0% | 100.0% | ||
a: n=43; b: Sensitivity=[TP/(TP+FN)]*100; c: Specificity=[TN/(TN+FP)]*100.
In contrast, Izzo et al. (2012) revealed that sensitivity of the Lateral flow test (LAT) compared to ELISA in detection of rotavirus was 67.8%, while specificity was high (95.2%). On the other hand, previous studies for the comparison of the rapid test kit with the RT-PCR revealed slightly higher sensitivity (83%) and specificity(100%) of the BRV (Saklı et al., 2019) while Klein et al. (2009) reported 71.9 % sensitivity and 95.3% specificity.
The application of molecular based assays can provide more information on the prevalence of BRV genotypes and can revolutionize information on the molecular basis of epidemiological variation among BRV (Badaracco et al., 2013). RT-PCR results compared to ELISA were relatively low (sensitivity; 71.4% and 100% specificity) (Table 3). Falcone et al. (1999) reported that RT-PCR was higher sensitivity due to its ability to detect one virus particle in fecal samples. In addition, stability of double‐stranded RNA of rotavirus may persist with pernicious conditions longer than viral antigen proteins. On the contrary, comparison of sensitivity and specificity for the available ELISA kits and qRT-PCR, previous findings have found very low sensitivity (44.7%) and specificity (86.4 %) (Izzo et al., 2012). The low sensitivity in this study can be attributed to the qRT-PCR reaction conditions and or inhibiting factors within the tested specimens especially with possibility for contaminant with bacteria and fungi from environment.
Twenty-nine samples were negative by using all tests, which suggests low number of virus particles (non-detectable) or the gastroenteritis due to another causative agent. Only two samples were found positive by ELISA that support the higher sensitivity of ELISA, while two samples were positive by both ELISA and rapid test while PCR was negative for those two samples. Also, five samples were positive by both ELISA and PCR however, were negative by rapid strip test which may be due to low virus particles that cannot be detected due to low sensitivity of rapid strip test.
Our results revealed that ELISA is an indispensable tool for diagnosis of BRV from clinical cases due to its simplicity, rapidity, low cost, and applicability without a need for trained personnel or expensive equipment.

Table 4: Percentage of nucleotides identity between BRoV_AHRI-1 Egypt strain and other Published reference strains on GenBank based on partial sequence of NSP5.
One sample representing to RT-PCR positive samples was sequenced to confirm identity of GPA BRV, the partial nucleotide sequence of the non-structural protein 5 (NSP5) of strain BRoV_AHRI_1 was submitted to the GenBank and has obtained the accession number MT210506. Analyzing the sequence data of the strain revealed that this strain has high identity percentage reached 100% with different strains (KX212877.1 TUR2015, KF697087.1 India 2012, JQ943572.1 BRA 2009 and Egyptian strain Menofia 2018), however, the similarity rate decreased slightly to 99.4% with other strains as LC133557.1 USA 1983, LC133546.1 THA 1989 and KF697089.1 India 2012 (Figure 2 and Table 4). Comparison for the sequence identity of the detected virus with recent isolates in Egypt and worldwide was carried out to assess the vaccination programs efficacy, matching identity with the recent Egyptian strains, which will help for updating the current vaccination protocol to be reliable and efficient against currently circulating BRV strains.
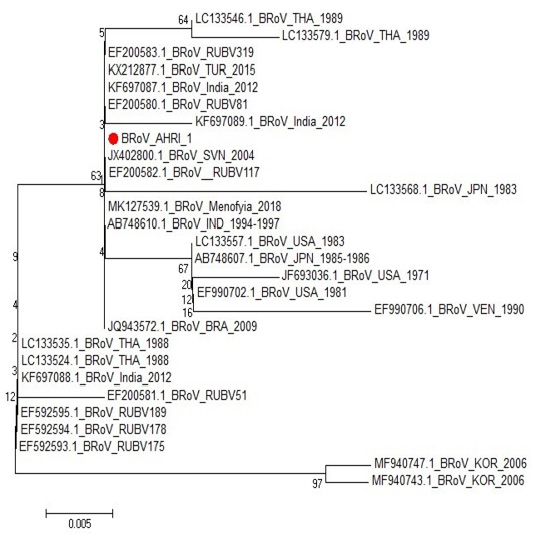
Figure 2: Phylogenetic tree for NSP5 partial nucleotide sequence of Bovine Rotavirus strain (BRoV_AHRI_1) (Acc. no MT210506) (Red circle) compared with other reference strains in GenBank database. The scale bar represents the number of substitutions per nucleotide.
CONCLUSIONS
Prevalence of bovine rotavirus in fecal samples collected from neonatal calves suggested its etiological role in neonatal calf diarrhea.
Management for BRV suspected clinical cases requires rapid and accurate diagnosis; therefore, using ELISA for detection of the viral antigen from fecal samples is a good alternative tool than both rapid test and RT-PCR. Likewise, further studies are required to develop more rapid, sensitive and specific assays for field diagnosis of BRV infection.
Author’s Contribution
First author conceived of the presented idea and collected samples, second and third authors developed the theory and the computations, carried out the experiment and wrote the manuscript.
All authors discussed the results and contributed to the final manuscript.
Conflict of interest
The authors have declared no conflict of interest.
REFERENCES





