Advances in Animal and Veterinary Sciences
Research Article
First Characterization of Class 1 Integron in Corynebacterium bovis Isolated from Subclinical Bovine Mastitis
Ashraf A. Abd El-Tawab1, Ashraf M. Ahmed2, Nabih A.M.3, Walaa H. Saad4*
1Department of Bacteriology, Immunology and Mycology, Faculty of Veterinary Medicine, Benha University, Egypt; 2Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Kafrelsheikh University, Kafr El-Sheikh 33516, Egypt; 3Animal Reproduction Research Institute (ARRI), Ministry of Agriculture, Cairo, Egypt; 4Central Diagnostic and Research Lab. of Fish, Poultry and Animal Diseases, Faculty of Veterinary Medicine, Kafrelsheikh University, Egypt.
Abstract | Integrons play major roles in spreading and expression of antibiotic resistance genes among bacteria especially Gram-negative bacteria. However, the incidences of class 1 integrons in Gram-positive bacteria remained elusive. The present study was conducted to monitor the prevalence of class 1 integrons in Corynebacterium bovis (C. bovis) isolated from cases of bovine mastitis in Egypt. A total of 35 Corynebacteria spp., isolated from milk samples of subclinical mastitis, were examined using PCR analysis with 16S rRNA specific primers. The results revealed a prevalence of 8 (22.8%) C. bovis strains in the examined samples. All isolates of C. bovis showed an elevated level of resistance, mostly against spectinomycin, streptomycin followed by tetracycline, ampicillin, kanamycin, chloramphenicol and sulfamethoxazole-trimethoprim. Using PCR and DNA sequencing, two isolates of C. bovis were identified containing class 1 integrons that encoded aminoglycoside adenyltransferase type A (aadA1) as a gene cassette. The aadA1 confers resistance to spectinomycin/streptomycin. Additionally, class 1 integron resistance genes in C. bovis isolates were described utilizing PCR and DNA sequencing. These resistance genes conferred resistance to several antimicrobial agents frequently used in dairy farming. This study reports, for the first time, the detection and identification of class 1 integron containing aadA1 gene in C. bovis of animal origin.
Keywords | PCR, DNA sequencing, aadA1, Gene cassette, C. bovis
Received | October 24, 2019; Accepted | February 17, 2020; Published | April 08, 2020
*Correspondence | Walaa H. Saad, Central Diagnostic and Research Lab. of Fish, Poultry and Animal Diseases, Faculty of Veterinary Medicine, Kafrelsheikh University, Egypt; Email: hamadasaad53@yahoo.com
Citation | El-Tawab AAA, Ahmed AM, Nabih AM, Saad WH (2020). First characterization of class 1 integron in Corynebacterium bovis isolated from subclinical bovine mastitis. Adv. Anim. Vet. Sci. 8(5): 452-457.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.5.452.457
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 El-Tawab et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Corynebacteria spp. are one of the most isolated pathogens responsible for subclinical mastitis in dairy cows. However, quick, reliable and simple techniques for the identification of species of the genus Corynebacterium are not currently available (Juliano et al., 2014).
The wide utilization or misuse of antibiotics in veterinary medicine initiates the development of resistance and increase incidence of multidrug resistance (MDR) in bacteria. This leads to implication of substantial public health impacts (Pyorala, 2009; Richet, 2012; Saini et al., 2012; Chen et al., 2017).
Integrons are portable hereditary components having a system of site-specific recombination, which permits capture the genes responsible for antibiotic resistance (Partridge et al., 2009; Chen et al., 2011). Integrons play important roles in spreading and expression of antibiotic resistance genes among bacterial populations through transposons or plasmids (Mazel, 2006). The integron gene sequence arrays are formed when multiple gene cassettes are integrated at one site on a plasmid or on the chromosome (Recchia and Hall, 1995, 1997).
According to the similarities of integrase amino acid sequence, integrons are classified into several classes; the most common one is class 1 integrons which are detected in Gram-negative bacterial isolates from various sources (Mazel, 2006; Chen et al., 2011; Mokracka et al., 2012). The class 1 integron contains two conserved segments as following; a 5’ conserved segment (5’-CS), a 3’ conserved segment (3’-CS) and in between them a recombination site at which the gene cassettes have been integrated (Collis et al., 2002; Correia et al., 2003). The 5’-CS made out of integration site (attI), an integrase gene (intI), promoters responsible for gene cassettes and the integrase expression (Ploy et al., 2000; Mazel, 2006). The 3’-CS involved sul1 genes and the truncated qacEΔ1, which confer resistance to sulfonamides and disinfectants, respectively (Paulsen et al., 1993).
The class 1 integrons genes are mostly detected in Gram-negative bacteria (Martinez-Freijo et al., 1998; Partridge et al., 2001). However, the incidence of class 1 integrons in Gram-positive bacteria remained least explored (Nesvera et al., 1998; Clark et al., 1999; Tauch et al., 2002).
The present study aim to detect C. bovis strains isolated from subclinical cases of bovine mastitis using PCR techniques and to genetically characterize class 1 integron resistance genes using advanced genomic approaches.
MATERIALS AND METHODS
Sampling and isolation
A total of 150 mastitis milk samples, obtained from cases of subclinical mastitis (positive for California mastitis test), were gathered from different farms of dairy cattle at Giza governorate in the Egypt. At these farms, cows suffered from decreased milk production with no clinical signs. A total of 35 isolates were identified as Corynebacteria species through microbiological cultures according to the method described by Oliver et al. (2004).
DNA extraction and PCR
The DNA extraction was carried out by utilizing multi-genomic DNA extraction kit (Qiagen, Germany), according to manufacturer’s instructions. All isolates of Corynebacteria spp. were subjected to PCR amplification (Table 2) using thermal cycler (Peltier Thermal Cycler - MG 960T enzyme® - USA) with the primer pairs (Sigma, USA) F-5′ GCGAACGGGTGAGTAACACG 3′ and R-5′ TCTGCGATTACTAGCGACTCCG 3′ (Huxley et al., 2004), as shown in Table 1.
Antimicrobial sensitivity test
The phenotypes of antimicrobial sensitivity of C. bovis isolates were tested on Mueller–Hinton agar (Oxoid, Basingstoke, Hampshire, United Kingdom) plates using a disk diffusion method according to the guidelines and standards showed by Clinical and Laboratory Standards Institute (CLSI, 2018). Antibiotic susceptibility of tested organisms were determined for tetracycline (TET), 30 µg; spectinomycin (SPX), 10µg; kanamycin (KAN), 30 μg; streptomycin (STR), 10 µg; sulfamethoxazole-trimethoprim (SXT), 23.75/1.25 µg and ampicillin (AMP), 10 μg. The sensitivity disks were punched from (Oxoid, UK). The recorded results were depending on the zone-size in chart of interpretation according to manufacturer’s instructions.
PCR and sequencing of class 1 integron from C. bovis
All PCR reactions were adjusted to 25 μl of total volume; 12.5 μl of 2xMaster Mix (Intron Biotechnology, Inc), 5 μl DNA of C. bovis, 1.25 μl of 20 pmol/μl) of each primers (Sigma, USA) of class 1 integron and finally, water nuclease-free up to 25 μL. All PCR reactions of C. bovis isolates were submitted to PCR amplification with thermal cycler (Peltier Thermal Cycler - MG 960T enzyme® - USA) with 5’-CS and 3’-CS primers specific to class 1 integron, as described in (Table 2) (Lin et al., 2016). This primer pair target the region between the 5’-conserved segment (5’-CS) and 3’-CS of class 1 integrons as previously described (Table 1).
Analysis of PCR products was carried out through gel electrophoresis (SCIE-PLAS, UK) on a 1% agarose gel prepared with Tris/Acetate/EDTA (TAE) (Intron Biotechnology, Inc) buffer plus agarose powder (Biotechnology grade AGA001.100 - Bioshop® Canada Inc) stained with ethidium bromide (0.5 μg/ml). 10-μL aliquot of a molecular marker containing a DNA Ladder (Intron Biotechnology, Inc) was added to the first well of each gel, control negative in the second, control positive (K. pneumoniae KPB 127 contain two integrons (dfrA12 - aadA2; and aadA2) isolated by (Hazim et al., 2016) in the third well and 10-μL of samples in the other wells (Figure 2) at 80 V for 1 h, after that single DNA bands were visualized by ultraviolet trans-illumination (UV) according to target size of gene as showed in Table 2. A QIA quick Gel Extraction Kit (Qiagen, Germany) was used for purification of PCR fragments from the agarose gel according to the manufacturer’s instructions. Finally, the sequencing for PCR product with both DNA strands was performed by using an ABI automatic DNA sequencer (Model 373; Perkin–Elmer).
Computer analysis of the sequence data
The data of DNA sequenced data of amplified gene was identified by the database of GenBank using the BLAST program available at the NCBI BLAST homepage (http://www.ncbi.nlm.nih.gov/BLAST/).
Table 1: Primers used for PCR reactions and DNA sequencing.
| Target | Primer name | Oligonucleotide sequence (5' - 3') | Amplicon size (bp) | Reference |
| C. bovis |
F R |
GCGAACGGGTGAGTAACACG TCTGCGATTACTAGCGACTCCG |
1250 bp |
Huxley et al. (2004) |
| Class I integron |
5′-CS 3′-CS |
GGCATCCAAGCAGCAAG AAGCAGACTTGACCTGA |
Variable |
Lin et al. (2016) |
Table 2: General PCR conditions of different primers.
| Gene | Pre denaturation | Denaturation | Annealing | Extension. | Cy. | Final extension | Target |
|
C. bovis: C-F C-R |
94 oC/2 min |
94 oC/30 sec |
58oC/30 sec |
72 oC/1 min |
30 | 72ºC/1 min | 1250 bp |
|
Class I integron 5′-CS 3′-CS |
94 oC/10 min |
94 oC/1 min |
55 oC/1 min |
72 oC/3 min |
30 | 72ºC/10 min | Variable |
Table 3: Incidence of C. bovis and class 1 integron.
|
Total no. of bacteriologically examined Corynebacteria spp. isolates |
C. bovis isolated by PCR | Class 1 integron isolatesby PCR | ||
| No. | % | No. | % | |
| 35 | 8 | 22.8 | 2 | 5.7 |
Percent according to total no. of Corynebacteria spp.
Table 4: Results of antimicrobial susceptibility patterns of C. bovis isolates and interpretation according to (CLSI. 2018).
| No of isolate | AMP | STR | SPX | KAN | SXT | CHL | TET |
| C2 | R | S | R | S | S | S | R |
| C3 | R | R | R | R | S | S | R |
| C6 | I | R | R | R | R | R | R |
| C8 | R | R | R | S | R | I | S |
| C11 | R | R | R | R | S | S | R |
| C16 | S | R | S | R | R | R | R |
| C23 | R | I | R | S | I | I | R |
| C31 | S | R | R | S | S | S | R |
| Incidence of resistance | 5 (62.5%) | 75% | 7 (87.5%) | 3 (37.5%) | 3 (37.5%) | 2 (25%) | 5 (62.5%) |
S: sensitive; R: resistant; I: intermediate; AMP, ampicillin; CHL, chloramphenicol; KAN, kanamycin; SPX; spectinomycin STR, streptomycin; SXT, sulfamethoxazole-trimethoprim; TET, tetracycline.
RESULTS
Bacteriological examination of collected 150 subclinical mastitis milk samples (positive for CMT), revealed 35 (23.3%) isolates of Corynebacteria spp. The antimicrobial sensitivity of the C. bovis to antimicrobial agents is shown in Table 2. The C. bovis isolates showed resistance against streptomycin (87.5%), spectinomycin (75%), tetracycline (62.5%), ampicillin (62.5%), kanamycin (37.5%), sulfamethoxazole-trimethoprim (37.5%) and chloramphenicol (25%). Class 1 integron resistance gene was found in 2 (5.7%) isolates, as shown in Table 4.
In the present study, 35 isolates of Corynebacteria spp. were inspected using PCR with 16S rRNA gene, which is a primer specific for C. bovis. The result showed 8 positive strains of C. bovis out of 35 (22.8%) tested samples (Table 3) with a target PCR amplicon size of 1250 bp (Figure 1, Table 1).
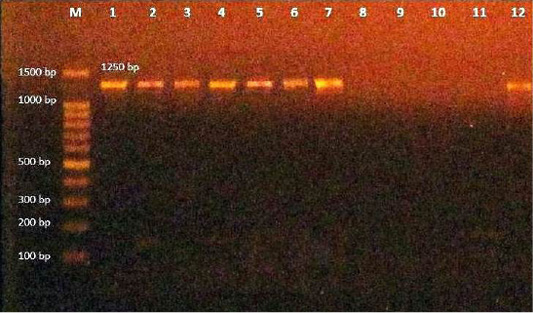
Figure 1: Amplification products of C. bovis was analyzed by electrophoresis on a 1.5% agarose gel. Lane M: 100-bp DNA ladder, lane (1-2-3-4-5-6-7-12)): positive at size 1250 bp. Lane (8-9-10-11): negative.
Further, PCR and DNA sequencing of 8 positive C. bovis fragments using primers specific to class 1 integrons revealed two isolates (5.7%) of C. bovis which have one type of class 1 integron with middle size (974 bp), as shown in (Figure 2) and contains a single gene cassette of aadA1 (aminoglycoside adenyltransferase type A), which confers resistance to streptomycin/spectinomycin. Up to date the aadA1 gene has not been previously identified in C. bovis.
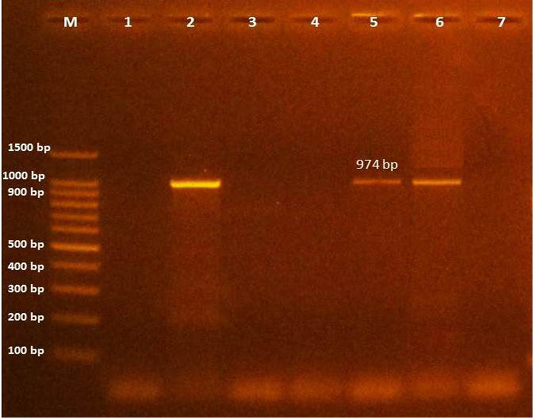
Figure 2: Amplification products of class 1 integron in C. bovis was analyzed by electrophoresis on a 1.5% agarose gel. Lane M: 100-bp DNA ladder, Lane (1): Control negative, lane (2): control positive (K.pneumoniae KPB 127 contain two integrons ( dfrA12 - aadA2; and aadA2), lane (3-4-7): negative, lane (5-6): positive at size 974 bp (aadA1).
DISCUSSION
In this study, 8 out of 35 isolates were identified as C. bovis using PCR with a faire percentage (22.8%). This rate is lower than 54.0% strains as C. bovis reported by Watts et al. (2000) and also Theel et al. (2012) which detects C. bovis with a percent 88.5%. Higher incidence of C. bovis was reported from sub-clinical mastitis (96.1% and 97.2%) by Juliano et al., 2014 and Huxely et al., 2004, respectively.
The class 1 integrons were first reported in 1989 (Stokes and Hall, 1989) and were described as reservoirs and an essential source of antimicrobial resistance genes within microbial populations (Fujimura et al., 2004; Xu et al., 2007, 2009). However, these studies have been conducted in Gram-negative bacteria with few exceptions (Tauch et al., 2002; Nandi et al., 2004). While in recent studies, class 1 integron had been observed as antibiotic resistance gene in Gram-positive bacteria, particularly in methicillin-resistant coagulase negative staphylococci (MRCNS) and methicillin-resistant Staphylococcus aureus (MRSA) (Xu et al., 2007, 2008, 2010).
Class 1 integron have been found mainly in Gram-negative bacteria (Mokracka et al., 2012; Malek et al., 2015). Up-to-date, only few reports on class 1 integron in Gram-positive bacteria are available. The previously published papers of class 1 integron in Gram-positive bacteria with gene cassettes are as following; (1) Class 1 integron containing aadA gene was first report in E. faecalis strain and confers resistance to streptomycin/spectinomycin (Clark et al., 1999). (2) The genome of some Gram-positive bacteria such as Staphylococcus aureus, coagulase-negative staphylococci, Staphylococcus warneri, Staphylococcus haemolyticus and Staphylococcus epidermidis carries class 1 integrons containing gene cassettes of dhfrXII-orfF-aadA2 which confers resistance to trimethoprim, unknown function protein and aminoglycoside adenyltransferase type A, respectively (Lei et al., 2006). (3) The aadA2 gene cassette on a 29-kb plasmid pCG4 in C. glutamicum, which confers resistance to streptomycin/spectinomycin (Nesívera et al., 1998). (4) Class 1 integrons containing aadA9 detected previously in C. glutamicum on a 27.8-kb R-plasmid pTET3, which confers resistance to spectinomycin, streptomycin and tetracycline (Tauch et al., 2002). (5) The aadA2 gene cassette in S. aureus, which conferred resistance to aminoglycoside (Xu et al., 2007). (6) Class 1 integron of different gene cassettes with 2 att sites and 3 arrays of cassettes were detected in methicillin-resistant S. aureus (MRSA) (Shengjie et al., 2011).
CONCLUSION
In the present study, class 1 integron was identified with gene cassette (aadA1) in two isolates of C. bovis (5.7 %) obtained from milk samples of subclinical mastitis cows using PCR and DNA sequencing. These resistance genes confer resistance to several antimicrobial agents frequently used in dairy farming. This study is the first report for detection and identification of class 1 integron containing aadA1 gene in C. bovis of animal origin.
ACKNOWLEDGEMENTS
This work was supported by Central Diagnostic and Research Lab. of Fish, Poultry and Animal Diseases, Faculty of Veterinary Medicine, Kafrelsheikh University, Egypt. We thank all staff members of Department of Bacteriology, Immunology and Mycology, Faculty of Veterinary Medicine, Benha University
Study limitation
Findings of this study were limited to a small sample size, also class 1 integron incidence is little in Gram positive bacteria. In addition, few Egyptian studies were accessed in this area of research.
Moreover, there were some difficulties related to the expense of chemicals and kits which restricted the application of PCR and DNA sequencing primers of multi drug resistance.
Authors Contribution
All authors contributed equally.
Conflict of interest
No conflict of interest declared.
REFERENCES





