Advances in Animal and Veterinary Sciences
Research Article
Molecular Determinants of Multiple Antibiotic Resistant E. coli Isolated from Food of Animal Origin
Fawzy Riyad El Seedy1, Ayman Amin Samy2, Eman A. Khairy2, Aya Atya Koraney2, Hala Sayed Hassan Salam1*
1Bacteriology, Mycology and Immunology Department, Faculty of Veterinary Medicine, Beni-Suef University, Beni-Suef, Egypt; 2Department of Microbiology and Immunology, National Research Center [NRC], Cairo, Egypt.
Abstract | Multiple antibiotic-resistant E. coli usually contaminates foods of animal origin representing a major threat to public health. The purpose of this study was to demonstrate multiple antibiotic-resistant E. coli as a serious food contaminating bacteria in milk, meat and their products. One hundred and twenty five (125) samples were collected from meat, milk and their products from different markets in Beni-Suef and Giza Governorates, Egypt. The isolated E. coli were subjected to antimicrobial sensitivity testing against ampicillin, tetracycline and streptomycin and resistance genes were detected using polymerase chain reaction (PCR). Out of 125 samples, 41 E. coli were isolated. Serogrouping of E. coli isolates revealed O157, O44, O119, O168, O6, O158, O164, O126 and O125 serogroups. Most of E. coli isolates were multidrug resistant (MDR). E. coli isolates were resistant to ampicillin, amoxicillin, kanamycin, tetracycline and streptomycin with percentages of 63.4, 61, 53.7, 41.5 and 14.6%, respectively. Out of tetracycline resistant isolates (n=17), 14 isolates harbored tetB gene (82.4%), while six isolates harbored tetA gene (35.3%). Meanwhile, both tetA and tetB were harbored in 4 isolates with a percentage of 23.5% Ampicillin resistance gene blaTEM found in 76.5% of the ampicillin resistant isolates. Streptomycin resistant genes aadA1 and aadA2 were represented in 50% and 0% of the isolates, respectively. Foods of animal origin is considered an important source of multiple antibiotic resistant E. coli that can be a major source of food-borne diseases.
Keywords | E. coli, MDR, Food of animal origin, Resistance genes, PCR
Received | November 20, 2019; Accepted | February 17, 2020; Published | March 20, 2020
*Correspondence | Hala Sayed Hassan Salam, Bacteriology, Mycology and Immunology Department, Faculty of Veterinary Medicine, Beni-Suef University, Beni-Suef, Egypt; Email: hala_saydh@yahoo.com
Citation | El-Seedy FR, Samy AA, Khairy EA, Koraney AA, Salam HSH (2020). Molecular determinants of multiple antibiotic resistant E. coli isolated from food of animal origin. Adv. Anim. Vet. Sci. 8(4): 347-353.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.4.347.353
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 El-Seedy et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Food-borne illness is one of the major public health problems all over the world. Foods of animal origin are considered serious sources of food-borne diseases worldwide. Moreover, the situation worsens in developing countries due to difficulties in achieving optimum hygienic food handling practices (Schlundt et al., 2004; Newell et al., 2010; Badi et al., 2018). Foodborne pathogens cause high economic losses in developing countries due to medical care and social costs (Fratmico et al., 2005). The food safety unit of WHO (World Health Organization) recorded that about 1.8 million children died from diarrhea caused by contaminated food and drinking water in 2005 (WHO, 2011). Antibiotics make revolution in the treatment of diseases. They are either used for prophylactic and/or therapeutic purposes in humans and animals. Some antibiotics are used as animal growth promoters. Unfortunately, similar antibiotics are being used in both veterinary and humans practice. The correlation between extensive use of antibiotics and the development of multiple antibiotic resistant bacteria is well authenticated (Garipcin and Seker, 2015; Zdolec, 2016; Jans et al., 2018). Due to the misuse of antimicrobials in animal production, multiple antibiotic resistant bacteria have been increasing over the last decades, in addition to the spread of the resistant foodborne zoonotic pathogens (Teuber, 2001; Zdolec, 2016). There is a great possibility for transmission of resistant genes to the food-related commensal bacteria or opportunistic pathogens which represents a potential hazard to consumers (Aarestrup et al., 2008) bacteria may represent an important threat to public health, as the antibiotic resistance genes can be transferred from resistant bacteria to other bacteria infecting humans. The acquisition and spread of resistance genes may be due to the misuse of antibacterial agents or through horizontal gene transfer (Threlfall et al., 2000; Chiu et al., 2002). Escherichia coli is recognized as an important pathogen of livestock such as cattle and poultry. It may cause severe humans diseases such as hemorrhagic colitis and hemolytic uremic syndrome; a serious complication may lead to death (Diercke et al., 2014; Bryan et al., 2015; Markland et al., 2015). Due to its ubiquity, the antibiotic resistant E. coli can transmit its resistance genes from other Enterobacteriaceae species in the environment (Pitout, 2013; Hooper and Jacoby, 2015). Therefore, E. coli could be used as a monitoring for the misuse of antibiotics either in food producing animals or in treatment of humans (Laarem et al., 2017). Many studies in recent years undertaken to assess the antibiotic resistance of bacteria in food of animal origin such as meat, raw milk, and their products (Bhoomika et al., 2016; Ranjbar et al., 2018). Molecular analysis of antibiotic-resistant mobile elements and antibiotic resistance genes indicates similar elements in bacterial isolates from both animals and humans, which explain the role of food of animal origin in the dissemination of resistant bacteria to humans through food chain (Teuber, 2001; Bhoomika et al., 2016).
Therefore, this study was designed to investigate the existence of multiple antibiotic-resistant E. coli and their molecular determinants in food of animal origin that collected from Beni-Suef and Giza Governorates markets.
MATERIAL AND METHODS
Samples
One hundred and twenty five (125) samples were randomly collected from milk, meat and their products from Giza and Beni-Suef Governorates markets (Table 1). All samples were aseptically collected in sterile plastic bags separately and transferred immediately under hygienic measures in ice box to the laboratory to be examined for presence of E. coli.
Isolation and identification of E.coli
The prepared samples were inoculated onto MacConkey agar plates (Oxoid). Pink colonies were picked up and streaked onto eosin methylene blue agar (Oxoid) to observe the metallic sheen appearance. Typical colonies were picked up onto nutrient agar and subjected to standard morphological and biochemical tests as well as serological identification of the identified isolates as described by Quinn et al. (2002).
Antimicrobial sensitivity testing for identified isolates
The antimicrobial sensitivity test was done by disk diffusion technique using antibiotic discs including ampicillin (10µg), amoxicillin (10µg), amoxicillin-clavulanate (20/10µg), streptomycin (10µg), kanamycin (30µg), ciprofloxacin (5µg), Trimethoprim- sulfamethoxazole (1.25/ 23.75 µg) and tetracycline (30µg) (Oxoid). The degree of sensitivity was interpreted according to the clinical laboratory standard institute (CLSI, 2017).
Genotypic detection of the resistance associated genes
Polymerase chain reaction was performed for detection of resistance genes in Biotechnology Center in the animal health institute according to Sambrook et al. (1989). The primers (Metabion, Germany) of target genes with their sequence and references are listed in Table 2.
RESULTS AND DISCUSSION
Isolation and identification of E. coli from different food samples
The results showed in Table 3 revealed that E. coli was totally isolated with a percentage of 32.8 arranged as 30.8% and 35% from samples of (raw milk and milk products) and (meat and meat products), respectively.
Nearly the same recovery rate of E. coli from meat and meat products was obtained by many studies worldwide (Ali et al., 2010; Gousia et al., 2011; Bhoomika et al., 2016). A higher prevalence rate was obtained by Abdaslam et al. (2014) who identified E. coli from meat products with a percentage of 50%. While a lower prevalence rate was obtained by Al-Zogibi et al. (2015) who recovered 17 E. coli isolates (11.33%) from raw meat samples. The same lower recovery rate of E. coli was found in meat with a percentage of 20% (Petternel et al., 2014). In the present work the incidence of E. coli isolated from milk and milk products is approximately the same obtained by many studies (Zeinhom and Abdel-Latef, 2014; Al-Zogibi et al., 2015). A higher recovery rate was recorded by many studies (Abike et al., 2015; Bhoomika et al., 2016).
Serogrouping of the identified isolates
Serogrouping of E. coli isolates revealed nine types while seven of the tested isolates were untyped with available sera. For each of the following serogroups (O6, O119, O125, O158, and O168) two isolates were identified with a percentage of 4.9, four isolates for every one of O 44 and O164 with a ratio of 9.77%. At the same time, O157 and O126 represented the highest prevalence (six isolates for each one; 14.6%). E. coli O157 which represented the highest prevalence in the study is a zoonotic food-borne
Table 1: Total number of product samples collected from sale markets.
| Total number of samples | Milk | Yoghurt | Kareesh | Minced meat | Burger | luncheon | ||||||
| 125 | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % |
| 28 | 22.4 | 18 | 14.4 | 19 | 15.2 | 20 | 16 | 20 | 16 | 20 | 16 | |
%: Percentages were calculated according to the total number of samples.
Table 2: Oligonucleotide primer sequences of target genes specific for E. coli.
| Target gene | Primer sequence (5′ → 3′) | Amplified product (bp) | References |
| TetA [A] | GGTTCACTCGAACGACGTCA | 576 bp | |
| CTGTCCGACAAGTTGCATGA | |||
| TetA [B] | CCTCAGCTTCTCAACGCGTG | 633 bp | |
| GCACCTTGCTCATGACTCTT | |||
|
BlaTEM |
ATCAGCAATAAACCAGC | 516 bp | |
| CCCCGAAGAACGTTTTC | |||
| AadA1 | TATCAGAGGTAGTTGGCGTCAT | 484 bp | |
| GTTCCATAGCGTTAAGGTTTCATT | |||
| AadA2 | TGTTGGTTACTGTGGCCGTA | 622 bp | |
| GATCTCGCCTTTCACAAAGC |
Table 3: Prevalence of E. coli isolated from the different food products during the study.
| Samples source | No. of examined samples | Recovered E. coli isolates | |
| Number positive | Percent (%) | ||
| Milk | 28 | 6 | 21.4 |
| Yogurt | 19 | 2 | 10.5 |
| Kareish cheese | 18 | 12 | 66.7 |
| Milk and milk products | 65 | 20 | 30.8 |
| Minced meat | 20 | 11 | 55 |
| Burger | 20 | 4 | 20 |
| Luncheon | 20 | 6 | 30 |
| Meat and meat products | 60 | 21 | 35 |
| Total | 125 | 41 | 32.8 |
%: Percentages were calculated according to the corresponding numberof examined samples.
Table 4: Antimicrobial susceptibility profile of E. coli isolates (n=41) in the examined samples during the study.
| Antimicrobial agents | Sensitive | Intermediate | Resistant | |||
| No. | % | No. | % | No. | % | |
| Penicillin group | ||||||
| Ampicillin | 13 | 31.7 | 2 | 4.9 | 26 | 63.4 |
| Amoxicillin | 13 | 31.7 | 3 | 7..3 | 25 | 61.0 |
|
β-lactam/β-lactamase inhibitor combinations |
||||||
| Amoxicillin-clavulanate | 37 | 90.2 | 2 | 4.9 | 2 | 4.9 |
| Aminoglycoside group | ||||||
| Streptomycin | 25 | 61.0 | 10 | 24.4 | 6 | 14.6 |
| Kanamycin | 16 | 39.0 | 3 | 7.3 | 22 | 53.7 |
| Tetracycline group | ||||||
| Tetracycline | 20 | 48.8 | 4 | 9.8 | 17 | 41.4 |
| Fluoroquinolones group | ||||||
| Enerofloxacin | 39 | 95.2 | 1 | 2.4 | 1 | 2.4 |
| Ciprofloxacin | 41 | 100 | 0 | - | 0 | - |
| Folate pathway inhibitor | ||||||
| Trimethoprim-sulfamethoxazole | 40 | 97.6 | 0 | 0 | 1 | 2.4 |
%: Percentages were calculated in relation to the total number of tested isolates.
pathogen of worldwide significance; elicits possibly life-threatening hemorrhagic colitis, hemolytic uremic syndrome and/or bloody diarrhea (Kanitpun et al., 2004). Previous studies in Brazil and Nigeria informed that most E. coli strains isolated from yoghurt raw milk and cheese belonged to enteropathogenic serogroups (Paneto et al., 2007; Abike et al., 2015). A former study in Egypt done by Ahmed and Shimamoto (2014) reported E. coli O157:H7 in cheese, chicken meat, beef and raw milk.
Antimicrobial susceptibility of the isolated E. coli
The results of E. coli sensitivity test (Table 4) revealed that most of the isolates were multidrug resistant (MDR). They were resistant to ampicillin, amoxicillin, kanamycin, tetracycline and streptomycin with percentages of 63.4%, 61%, 53.7%, 41.5% and 14.6%, respectively. On the other hand, high sensitivity pattern was observed to ciprofloxacin (100%), trimethoprim-sulfamethoxazole (97.6%), enerofloxacin (95.2%) and amoxicillin-clavulanate (90.2%). MDR was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories (Magiorakos et al., 2012). Similar MDR E. coli isolates were recovered by many recent investigations (Hinthong et al., 2017; Lahuerta-Marin et al., 2017; Martínez-Vázquez et al., 2018; Ranjbar et al., 2018). Nearly the same findings of resistant E. coli against ampicillin, amoxicillin, kanamycin, tetracycline and streptomycin were obtained by many studies (Lei et al., 2010; Rashid et al., 2013; Rasheed et al., 2014; Laarem et al., 2017).
Genotypic detection of resistance associated gene
The accurate and rapid detection of antibiotic resistance genes is extremely important in preventing the spread of infections. PCR-based molecular technique is preferred for detection of the antibiotic resistance genes. It is a rapid, specific, and accurate method for detection of bacteria with resistant genes of interest (Hinthong et al., 2017; Lahuerta-Marin et al., 2017; Martínez-Vázquez et al., 2018; Ranjbar et al., 2018).
Amplification tetA and tetB (genes in tetracycline resistant isolates)
PCR was carried for amplification of tetA (576 bp) and tetB (633 bp) genes in tetracycline resistant isolates. Out of 17 tetracycline resistant isolates, 14 isolates harbored tetB gene (82.4%)), while 6 isolates harbored tetA gene (35.3%). Meanwhile, both tetA and tetB were harbored in 4 isolates with a percentage of 23.5% (Figures 1 and 2).
Amplification of blaTEM gene in ampicillin resistant isolates
Antimicrobial susceptibility revealed that 26 isolates were resistant to ampicillin, a total of 17 isolates were selected for genotypic detection of blaTEM gene (516 bp), about 76.5% (13/17 isolates) harbored blaTEM gene (Figure 3).

Figure 1: Lanes 1, 6, 8, 13, 15, 16: positive amplification of tetA gene at 576 bp Pos.: positive control; Neg.: negative control; M: DNA ladder.
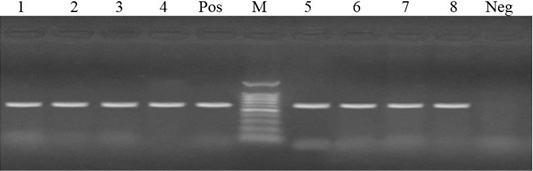
Figure 2: Lanes 1-8: positive amplification of tetB gene at 633 bp. Neg: negative control; pos: positive control; M: DNA ladder.
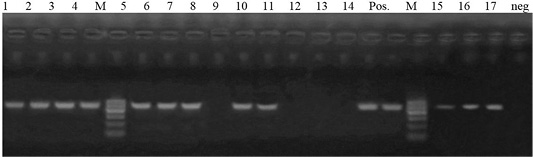
Figure 3: Lane 1-7, 9, 10, 14, 15- 17: positive amplification of blaTEM gene at 516pb fragment. Neg: negative control; pos: positive control; M: DNA ladder.
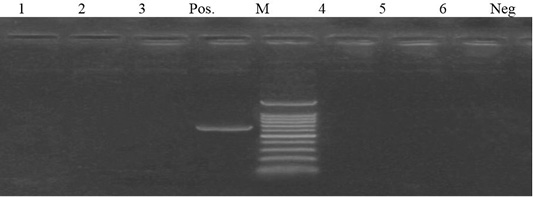
Figure 4: Lane 1- 6 negative amplification of aadA2 gene at 622 bp. Neg: negative control; pos: positive control; M: DNA ladder.
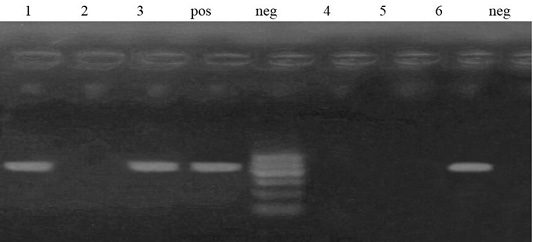
Figure 5: Lane 1, 3, 6 positive amplification of aadA1 gene at 484 bp. Neg: negative control; pos: positive control; M: DNA ladder.
Amplification of aadA1 and aadA2 genes in streptomycin resistance isolates.
Out of 6 streptomycin resistant isolates, aadA1and aadA2 genes were detected in a percentage of 50% (3 isolates) and 0%, respectively (Figures 4 and 5).
Many researchers identified resistant genes among E. coli isolates (Sunde and Norstrom, 2006) and reported that aadA1, blaTEM, tetA and tetB genes are responsible for streptomycin, ampicillin, and tetracycline resistance.
Multi-drug resistance was recognized in 32% of the E. coli isolates by Guerra et al. (2003). The most prevalent resistances were detected against tetracycline, sulfamethoxazole, ampicillin, streptomycin, and spectinomycin, the predominant resistance genes were: ampicillin, blaTEM 1-like (92%), streptomycin, aadA1 (61%), tetracycline, tetA (66%) and tetB (42%). One-third of the E. coli isolates recovered by Jouini et al. (2009) had MDR phenotypes. The highest resistance was detected against ampicillin, tetracycline and streptomycin. Different variants of blaTEM, (tet - aadA), and aac (3) genes were recovered from most E. coli isolates resistant to ampicillin, tetracycline and streptomycin, respectively. High-resistance profile was observed in most E. coli isolated by Martínez- Vázquez et al. (2018), about 92.4% of the isolates showed a multi-drug resistant phenotype with resistance to ampicillin (92%) and tetracycline (75%), tetA and tetB were detected in 56% of isolates while aadA in 17%. Recent study concluded that the highest prevalence of resistant E. coli isolated from milk and dairy products was against ampicillin (100%), gentamicin (100%) and tetracycline (96.87%), the most commonly detected antibiotic resistance genes was tetA gene (76.56%) (Ranjbar et al., 2018).
CONCLUSIONS
Based on the previous results, it was concluded that milk, meat and their products may play a role in the spread of MDR E. coli, which is considered a reservoir of many antibiotic resistance genes that can be conveyed to other pathogenic bacteria representing a potential public health threat.
Authors Contribution
All authors contributed equally.
Conflict of interest
The authors declare that they have no conflict of interest.
REFERENCES





