Advances in Animal and Veterinary Sciences
Research Article
Mycological Identification of Some Fungi Isolated from Meat Products and Spices with Molecular Identification of Some Penicillium Isolates
Ashraf A. Abd El-Tawab1, Eman M. El-Diasty2, Dalia F. Khater3, Yahya M. Al-baaly3*
1Department of Bacteriology, Immunology and Mycology, Faculty of Veterinary Medicine, Benha University, Egypt; 2Department of Mycology, Animal Health Research Institute (AHRI), Ministry of Agriculture, Dokki, Giza, Egypt; 3Tanta lab branch, Animal Health Research Institute (AHRI), Ministry of Agriculture.
Abstract | This study was done to determine the fungal quality of meats products (luncheon, basterma, kofta and burger) and spices (used in meat processing) marketed in some groceries and supermarkets in Gharbia Governorate under various trade names and molecular (ITS) characterization for some Penicillium species. In 120 tested meat products and 33 spices samples, 9 mould genera were detected. The identified mould genera were Aspergillus, Penicillium, Acremonium, Cladosporium, Geotrichum, Mucor, Claveolaria, Emericella Nudulans and Scorulopsis. The main identified Penicillium species were; P. citrinum, P. aurantigreum P. chrysogenum were the most predominant species isolated from burger samples, while from kofta samples, P. Paxilli and P. restrictum were the most predominant isolated species. P. citreognigrum and P. carneum only detected in luncheon and basterma samples. In spices the main identified Penicillium species were; P. aurantigreum, P. carneum, P. chrysogenum, P. citreognigrum, P. citrinum, P. crustosum, P. Paxilli, P. restrictum, P. implicatum and P.simplicissimum. Three Penicillium isolates (P. chrysogenum, P. carneum and P. restrictum) from meat products and one Penicillium isolate (P. crustosum) from spices were subjected to PCR identification, and were sequenced in both directions. The obtained sequences were deposited under (Gene bank accession number: MK643347 P. restrictum isolate AYMA4, MK643348 P. chrysogenum isolate AYMA5, MK643349 P. carneum isolate AYMA6 and MK643350 P. crustosum isolate AYMA7. Sequences of Penicillium isolates were analysed with phylogenetic tree and percent identity. Phylogenetic analysis of Penicillium isolates using ITS sequence data grouped them into four clusters. PCR and gene sequence are easy and fast diagnostic methods. As conclusion meat products and spices highly contaminated with mould especially Aspergillus spp and Penicillium spp which may gain access during the manufacturing process leading to high economic losses and have a public health hazard due to the production of mycotoxins.
Keywords | Meat products, Spices, Moulds, Penicillium, PCR
Received | November 10, 2019; Accepted | December 26, 2019; Published | January 18, 2020
*Correspondence | Yahya M. Al-baaly, Tanta lab branch, Animal Health Research Institute (AHRI), Ministry of Agriculture; Email: yahyamustafa200@gmail.com
Citation | Abd El-Tawab AA, El-Diasty EM, Khater DF, Al-baaly YM (2020). Mycological identification of some fungi isolated from meat products and spices with molecular identification of some penicillium isolates. Adv. Anim. Vet. Sci. 8(2): 124-129.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.2.124.129
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Abd El-Tawab et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Meat products are the most palatable of highly nutritive value foods for human being as they are important sources for protein, fat, essential amino acids, minerals, vitamins and other nutrients (Biesalski, 2005). On the other hand, they are considered as an ideal culture medium for growth of many organisms because of the high moisture, the high percentage of nitrogenous compounds, plentiful supply of minerals, some fermentable carbohydrates (glycogen) and of a favorable pH for most microorganisms (Alahakoon et al., 2015).
Moulds are common inhabitants of soil, water and may be dispersed through the air and water and by the activities of small animals, particularly insects (Rawat, 2015). Mould gaining access into meat product from meat, spices, and other ingredients, from environment, equipment, and handlers during processing affect the status of the products (Morshdy et al., 2015). Contamination of meat products with different mould species considers a real hazard as it affecting the quality of these meat products by increasing the opportunity for its spoilage and deterioration (Alcaide-Molina et al., 2009).
The increase in the production of processed foods and high demand for meat is the major reason behind the rapid increase in spices consumption (Little et al., 2003). Spices are largely produced in countries where tropical climates are favorable to mycotoxin contamination. Furthermore, they are usually dried on the ground in the open air in poor hygienic conditions that even more promote growth of mold and production of mycotoxin (Martins et al., 2001).
The growth of some mould species is dangerous, because they may be produced mycotoxins (Empting, 2009). Mycotoxin; is one of the problems threatening human and animal health in food industry (Kaynarca et al., 2019). Studies have discussed their toxigenic, nephrotoxic, hepatotoxic, carcinogenic, immunosuppressive and mutagenic characteristics of mycotoxins (Da Rocha et al., 2014).
Molecular approaches have been applied as an alternative assay replacing cumbersome and time-consuming microbiological and chemical methods for the detection and identification of mould (Niessen, 2008). PCR-based methods for simultaneous detection of mould are useful tools to be used in food safety programs. These rapid and sensitive techniques allow taking corrective actions during food processing or storage for avoiding accumulation of mycotoxins in them (Rodríguez et al., 2017). Internal transcribed region of the DNA (ITS) are the best tool for the identification of fungi (Mirhendi et al., 2007). Therefore, this study was carried out to throw light on the mycological aspects of some meat products and spices with molecular characterisation of some Penicillium isolates existing in such products.
MATERIALS AND METHODS
Isolation and Identification of mould
Twenty five grams from each sample of 120 meat products and 33 spices (collected from supermarkets in Gharbia governorate, Egypt) were carefully and aseptically homogenized in a blender with 225 ml of sterile peptone water 0.1% to form a dilution of 1:10, from each tenth fold serial dilutions were accomplished up to 106. One ml of the previously prepared serial dilution was separately poured into duplicated Petri dishes carefully and mixed with Dichloran Rose Bengal Chloramphenicol (DRBC) agar (Oxoid) then incubated at 250C for 5-7 days. The isolated colonies were picked up from the DRBC agar and cultured into slope Sabauroud dextrose agar (SDA) (Oxoid) and incubated at 250c for 5-7 days after then transferred to Malt Extract Agar (MEA) (Oxoid) and Czapek yeast agar (CYA) (Oxoid) plates for identification. Colony appearance, exudate production, pigmentation and reverse coloration were assessed and colony diameters were measured and recorded (Pitt and Hocking, 2009).
PCR amplification and sequencing
DNA extraction from four penicillium isolates using DNeasy plant Mini kit Qiagen Genomic as described by manufacturer manual of Qiagen, Germany. Cat. No. 69104. DNA samples were tested in 50 μl reaction volume in a 0.2 ml eppendorf tube, containing 25 μl PCR Masster Mix, 1 μl of each primer, 3 μl target DNA, complete to a final volume of 50 μl with sterile PCR water. Penicillium isolates were identified via ITS-PCR of rDNA region with ITS15´-TCCGTAGGTGAACCTGCGG-3´ and ITS25´-TCCTCCGCTTATTGATATGC-3´ (560 bp) (White et al., 1990). PCR amplification conditions for four Penicillium isolates were: 5 min initial denaturation step followed by 35 cycles at 95 °C for 30 sec, 35 cycles at 560C for 30 sec and 35 cycles for 72 °C for 1min and a final extension step at 72 °C for 10 min. Amplification products were electrophoresed in agarose gels (1.5% w/v) (Agarose, Sigma, USA) and stained with ethidium bromide using Gene Ruler 100bp DNA Ladder (H3 RTU, Cat. No. DM003-R500 Gene Direx, BIO-HELIX Co., LTD). The amplified fragments were purified using Gene Jet PCR purification kit; Fermentas (Cat no. KO701). Sequencing was performed by sequencing to the PCR product on GATC Company by use ABI 3730x1 DNA sequences by using forward and reverse primers. Sequences were deposited at gene bank and phylogenetic analysis was done by MEGA X (Kumar et al., 2018), while sequence divergence and identity percent by DNA star (Felsenstein, 1985) and (Tamura et al., 2004).
RESULTS
Aspergillus spp. was the most prevalent species in the examined samples. It constituted 24 (88.88%) in luncheon, 36 (85.71%) in kofta, 35 (85.36%) in basterma and 46 (58.23%) in burger followed by Penicillium spp. 3 (11.12%), 6 (14.29%), 3 (7.32%) and 12 (15.18%) respectively. Mucor spp.3 (7.32%) in basterma, 9 (11.42%) in burger, while Geotrichum spp. 6 (7.59%), Cladosporium spp, Acremonium spp. 3 (3.79%) detected in burger samples only. While in spices, the predominant mould genera isolated were Aspergillus spp. 86 (55.48%), Penicillium 24 (15.48%), Mucor spp 21 (13.55%), Cladosporium spp 9 (5.80%), Acremonium spp 3 (1.94%) Scorulopsis 6 (3.85%), Claveolaria 3 (1.94%) and Emericella.nudulans 3 (1.94%). Further identification to Penicillium isolates was done in meat products, the predominant Penicillium species isolated from burger samples were P. citrinum 6 (7.59%) followed by P. aurantigreum and P. chrysogenum 3 (3.795%). Where P. Paxilli and P. restrictum were the most isolated Penicillium species from kofta samples with incidence rate 3 (7.14%)
Table 1: Incidence of mould isolated from examined meat product samples.
| Isolated mould | Luncheon | Kofta | Basterma | Burger | Spices | |||||
| No | % | No | % | No | % | No | % | No | % | |
|
Aspergillus spp. |
24 | 88.88 | 36 | 85.71 | 35 | 85.36 | 46 | 58.23 | 86 | 55.48 |
|
Acremonium spp. |
- | - | - | - | - | - | 3 | 3.79 | 3 | 1.94 |
|
Cladosporium spp. |
- | - | - | - | - | - | 3 | 3.79 | 9 | 5.80 |
|
Geotrichum spp. |
- | - | - | - | - | - | 6 | 7.59 | - | - |
|
Penicillium spp. |
||||||||||
| P. aurantigreum | - | - | - | - | - | - | 3 | 3.795 | 6 | 3.87 |
| P. carneum | - | - | - | - | 3 | 7.32 | - | - | - | - |
| P. chrysogenum | - | - | - | - | - | - | 3 | 3.795 | 3 | 1.94 |
| P. citreognigrum | 3 | 11.12 | - | - | - | - | - | - | - | - |
| P. citrinum | - | - | - | - | - | - | 6 | 7.59 | 3 | 1.94 |
| P. crustosum | - | - | - | - | - | - | - | - | 6 | 3.87 |
| P. Paxilli | - | - | 3 | 7.145 | - | - | - | - | - | - |
| P. restrictum | - | - | 3 | 7.145 | - | - | - | - | - | - |
| P.implicatum | - | - | - | - | - | - | - | - | 3 | 1.94 |
| P.simplicissimum | - | - | - | - | - | - | - | - | 3 | 1.94 |
|
Total of Penicillium spp |
3 | 11.12 | 6 | 14.29 | 3 | 7.32 | 12 | 15.18 | 24 | 15.48 |
|
Mucor spp |
- | - | - | - | 3 | 7.32 | 9 | 11.42 | 21 | 13.55 |
| Claveolaria | - | - | - | - | - | - | - | - | 3 | 1.94 |
| Emericella. Nudulans | - | - | - | - | - | - | - | - | 3 | 1.94 |
| Scorulopsis | - | - | - | - | - | - | - | - | 6 | 3.85 |
| Total | 27 | 100 | 42 | 100 | 41 | 100 | 79 | 100 | 155 | 100 |
for both isolates. P. citreognigrum 3 (11.12%) isolated from luncheon While P. carneum 3 (7.32%) from basterma, While in spices the isolated Penicillium species were 6 (3.87%), 3(1.94%), 3 (1.94%), 6 (3.87%), 3 (1.94%), 3 (1.94%) for P. aurantigreum, P. chrysogenum, P. citrinum, P. crustosum, P. implicatum, P. simplicssimum, respectively. (Table 1).
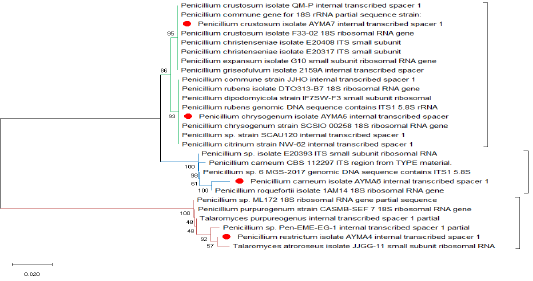
Three Penicillium isolates (P. chrysogenum, P. carneum and P. restrictum) from meat products and one Penicillium isolate (P. crustosum) from spices were positive on agarose gel electrophoresis (Figure 1) and the strains were successfully amplified and sequenced. The obtained sequences were deposited at NCBI under accession No. (Gene bank accession number: MK643347 P. restrictum isolate AYMA4, MK643348 P. chrysogenum isolate AYMA5, MK643349 P. carneum isolate AYMA6 and MK643350 P. crustosum isolate AYMA7. Penicillium isolates compared with published strains in Gene Bank through phylogenetic tree (Figure 2) and percent identity (Figure 3).
DISCUSSION
The presence of moulds may cause spoilage of meat products by breaking down their components and liberating different acids and gas with subsequent change of their odour and flavor. Some moulds are capable of producing toxic metabolites known as mycotoxins such as aflatoxins which may cause carcinogenic effect (Pitt and Hocking, 2009).
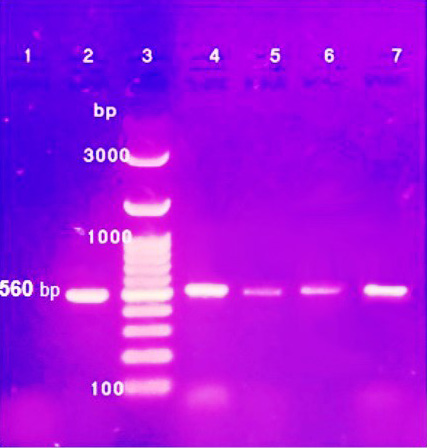
Figure 1: PCR products of ITS1-ITS2 regions from representative clinical isolates of Penicillin species Lane: 1 control Negative; Lane 2: control positive; Lane 3: 100 bp DNA ladder; Lane 4: P. restrictum; Lane 5: P. chrysogenum; Lane 6: P. carneum and Lane 7: P. crustosum.
In recent years, there has also been a steady increase in the production and consumption of processed meat products worldwide because of their high nutritive value and convenience (Rajic et al., 2007).
The results of mould incidence in meat products (Table 1) agreed with the results reported by (Shaltout and Salem, 2000; Brr et al., 2004; Zohri et al., 2014; Morshdy et al., 2015) who determined that Aspergillus spp. and Penicillium spp. were the most predominant mould species isolated from meat products samples. Occurrence of various types of molds in meat products may be attributed to the condition of the environment at which the meat products prepared, beside manufacturing rooms, stores, refrigerators, which are very suitable for the development of mold on meat and inside the meat products.
Spices have been used in many industries. They are commonly heavily contaminated with moulds. The most frequent fungal contaminants of spices are species from the genera Aspergillus and Penicillium. (Kocic´-Tanackov et al., 2007). Data presented in Table 1 revealed the dominant of Aspergillus and Penicillium spp. in all examined spices samples was in correct manner with the results of (Mandeel, 2005; Farghaly, 2006; Bokhari, 2007; Hashem and Alamri, 2010) who reported that Aspergillus and Penicillium spp. were the main components of spices.
The large numbers of species which constitute the genus Penicillium occupy a wide spectrum of habitats in our environment. As a consequence, many have become economically important in either harmful or useful roles. Some species cause deterioration of wide range of stored products (Frisvad and Samson, 2004). Some species of genus Penicillium might induce pulmonary infection, external otomycosis, mycotic keratitis and endocarditis (Birera et al., 1998).
Identification of Penicillium species is not easy. Many common species look like to the uninitiated. At the same time, there is a great deal of variability within the species. In recent years, molecular approaches have been used increasingly in identification and phylogenetic classification of filamentous fungi and their application has led to the reconsideration of several genera (Bruns et al., 1991).
Three Penicillium isolates (P. chrysogenum, P. carneum and P. restrictum) from meat products were examined by molecular methods polymerase chain reaction (PCR) with using ITS primer. PCR products of isolated strains were positive on agarose gel electrophoresis of PCR amplification products (Figure 1). Internal Transcribed Spacer (ITS) regions of fungal ribosomal DNA (rDNA) are highly variable sequences of great importance in distinguishing fungal species by PCR analysis (Martin and Rygiewicz, 2005). In the genus Penicillium, the region spanning of the nuclear ribosomal internal transcribed spacer has been investigated to clarify the subdivision within the genus and to evaluate phylogenetic relationship of some species. However, the ribosomal DNA gene has too few informative differences to reveal the phylogeny of Penicillium (Samson et al., 2004).
Phylogenetic analysis showed that P. restrictum isolate AYMA4 accession no. (MK643347) was isolated but in different branches, away from P. chrysogenum isolate AYMA5, P. carneum isolate AYMA6, P. crustosum isolate AYMA7 but remained in the same cluster. While P. restrictum isolate AYMA4 accession no. (MK643347) still in the same group of Talaromyces atroroseus isolate JJGG-11 and Penicillium sp. Pen-EME-EG-1. While P. chrysogenum isolate AYMA5 and P. crustosum isolate AYMA7 had common ancestor. P. chrysogenum isolate AYMA5 still in the same group of Penicillium chrysogenum strain SCSIO 00258 and Penicillium rubens isolate DTO313-B7. P. carneum isolate AYMA6 was isolated but in different branches, away from P. chrysogenum isolate AYMA5 and P. crustosum isolate AYMA7 but remained in the same cluster and still in the same group of Penicillium roquefortii1AM14 (Figure 2).
The sequence similarity of P. restrictum isolate AYMA4 for the strains of P. chrysogenum isolate AYMA5, P. carneum isolate AYMA6, P. crustosum isolate AYMA7, Talaromyces purpureogenus, Talaromyces atroroseus isolate JJGG-11 and Penicillium sp. Pen-EME-EG-1 were 80.1%, 78.2%, 79.8%, 99.2%, 99% and 99%. while the sequence similarity of P. chrysogenum isolate AYMA5 for P. carneum isolate AYMA6 and P. crustosum isolate AYMA7, Penicillium chrysogenum strain SCSIO 00258 and Penicillium rubens isolate DTO313-B7 were 94.7%, 98.7%, 100% and 99.6%. On the other hand, the sequence similarity of P. carneum isolate AYMA6 for P. crustosum isolate AYMA7and Penicillium roquefortii isolate 1AM14 were 95.1% and 99%. While the sequence similarity of P. crustosum isolate AYMA7 for Penicillium crustosum isolate F33-02 18S, Penicillium christenseniae isolate E20408, Penicillium expansum isolate G10, Penicillium griseofulvum isolate 2159A were 100% (Figure 3).
CONCLUSION
Meat products and spices highly contaminated with Aspergillus spp. and Penicillium spp. which may gain access during the manufacturing process leading to high economic losses and have a public health hazard due to the production of mycotoxins. The result demonstrates the fact that the unhygienic and poor sanitary conditions under which meat products and spices are handled and processed are not acceptable from sanitary point of view. It has further evidence that the undesirable level of contamination which might have acquired from the environment and agents. and to obtain wholesome safe and sound meat products, the principles Good Manufacturing Practices (GMP) and Hazard Analysis and Critical Control Point (HACCP) must be adopted.
ACKNOWLEDGEMENTS
This work was supported by Department of Mycology, Animal Health Research Institute (AHRI), Egypt. We thank all staff members of Department of Bacteriology, Immunology and Mycology, Faculty of Veterinary Medicine, Benha University
Authors Contribution
All authors contributed equally.
CONFLICT OF INTEREST
No conflict of interest declared.
References