Advances in Animal and Veterinary Sciences
Research Article
Molecular Characterization of Listeria Monocytogenes Isolated from Different Animal-Origin Food Items from Urban and Rural Areas
Azza S. El-Demerdash1* and Mona T. Raslan2
1Animal Health Research Institute, Zagazig, Egypt; 2Animal Health Research Institute, Dokki, Egypt
Abstract | Listeriosis is classified as the third main cause of food-borne diseases and it is one of the significant bacterial zoonotic infections causing high fatality rates worldwide. The aim of this research was to isolate and characterize Listeria monocytogenes (L. monocytogenes) from different food items in Sharkia Province, Egypt. A total of 200 food samples were collected aseptically from local markets and subjected to the phenotypic and genotypic characterization of L. monocytogenes. Listeria species were isolated from 28 (56%), 9 (18%), 3 (6%) and 5 (10%) of the minced meat, poultry meat, tilapia fish, and raw milk samples, respectively. Overall, L. monocytogenes were isolated from 12 and 8 samples from urban and rural areas with percentages of 6 and 4%, respectively. Of interest, inlA, actA, prfA, and hlyA virulence genes were detected in all 20 L. monocytogenes isolates by multiplex PCR. In conclusion, L. monocytogenes is an organism of public health implications, and its recovery from the food samples sold at retail outlets indicates a breach of quality assurance.
Keywords | L. monocytogenes, Food, Multiplex PCR, Virulence genes
Received | September 11, 2019; Accepted | October 06, 2019; Published | October 10, 2019
*Correspondence | Azza S. El-Demerdash, Animal Health Research Institute, Zagazig, Egypt; Email: Dr.azzasalah@yahoo.com
Citation | El-Demerdash AS and Raslan MT (2019). Molecular characterization of listeria monocytogenes isolated from different animal-origin food items from urban and rural areas. Adv. Anim. Vet. Sci. 7(s2): 51-56.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.s2.51.56
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 El-Demerdash et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Listeria monocytogenes (L. monocytogenes) is an opportunistic microorganism which causes extreme food-borne infirmity, commonly known as Listeriosis (Gray et al., 2006). It is ranked as one of the major causative agents for high mortality rates among all foodborne bacterial infections. The instance of listeriosis has been sporadic in African countries. Recently, there has been an expansion in the prevalence of human listeriosis, comprising food contamination in Egypt which iteratively detected in several dairy products, ready-to-eat foods, fish and fish products (Ismaiel et al., 2014; Kamar et al., 2014; Mohamed et al., 2016).
L. monocytogenes is a rod-shaped Gram-positive bacterium that is motile and non-spore-forming. It belongs to the facultative intracellular bacteria that attack, replicate and multiply in an assortment of cells through specific mechanism (Vasconcelos et al., 2008). After ingestion of contaminated food with L. monocytogenes, it crosses the intestinal barrier to the mesenteric lymph nodes. The bacteria disseminate via the bloodstream to the liver and spleen where they multiply. The bacteria are now able to reach the brain and, in case of pregnant women, also the placenta and the fetus. Entry into host cells is mediated by the internalins (Inl). Inside the cell, the bacterium is surrounded by a vacuole from which it rapidly escapes via listeriolysin O (LLO) by the action of hlyA gene (Churchill et al., 2006). Then, L. monocytogenes replicates in the cytoplasm and assembles an actin tail at the posterior pole of the bacterium where the surface protein (actA) is located. The actin tail is used to move in the cytoplasm and to spread into the neighboring cells via membrane protrusions (Vázquez-Boland et al., 2001; Liu et al., 2007a; Aurora et al., 2008; Piana et al., 2012).
Expression of the virulence genes mentioned above requires a transcriptional activator called prfA which is encoded by a gene in the same genomic cluster (Kathariou, 2002).
Therefore, dependable recognition of pathogenic L. monocytogenes has been proposed to be perfectly established upon the disclosure of virulence indicators (InlA, hlyA, actA, and prfA), but the determination of single virulence gene by uniplex PCR is not constantly adequate to recognize pathogenic L. monocytogenes (Di Ciccio et al., 2012; Jadhav et al., 2012). Moreover, the presence of spontaneous mutations may lead to the absence of one or more specific virulence determinants for some L. monocytogenes strains (Soni et al., 2010; Clayton et al., 2011; Di Ciccio et al., 2012).
In this way, the simultaneous discovery of a virulent pattern in one step minimizes the working period and will be utilized in a huge scanning for distinguishing the pathogenic listeria isolates. Multiplex PCR encompassing determination of these virulence genes has advanced as a zone of extraordinary potency (Chen and Knabel, 2007; Kérouanton et al., 2010; Shen et al., 2013).
Taking into consideration all these aspects, the present study investigated the prevalence of L. monocytogenes in animal-origin food items from different sources and areas in addition to determining some virulence determinants of isolates.
MATERIALS AND METHODS
Sampling
A total of 200 samples from different sources (minced meat, poultry meat, tilapia fish and raw milk) - 50 each- were collected from different urban and rural areas -100 each- in Sharkia province, Egypt.
Isolation and identification of L. monocytogenes
The International Organization for Standardization methods were used for isolation and identification of L. monocytogenes from different food specimens ( ISO 11290-1: 1996/Amd 1 2004) by the aid of enrichment broths as half Fraser and full Fraser; Oxford and Agar Listeria Ottaviani and Agosti (ALOA) agar. The purification was achieved onto Tryptone Soya Yeast Extract agar (TSYEA). Pinpoint colonies presumptively suspected as listeria isolates on TSYEA plates were sequentially submitted to comprehensive analyses including beta hemolysis on sheep blood agar and some biochemical tests such as sugar fermentation on rhamnose and xylose broths, reactions on triple sugar iron agar and oxidase (Hitchins, 2003).
Molecular investigation and characterization
The methodology of QIAGEN DNeasy was utilized to extract the DNA of the L. monocytogenes isolates with a minor adjustment in centrifugation parameters that is multiplying the centrifugation time. Then, PCR amplification was achieved with a PTC-100 programmable thermal cycler (Peltier-Effect cycling, MJ, UK) with adjusting the final volume of the reaction mixture to 50 µL consisting of 25 µL of DreamTaq TM Green Master Mix (2X) (Fermentas, USA), 0.2 µL of 100 pmole of each primer (Sigma, USA), 5 µLof template DNA and water nuclease-free up to 50 µL.
The multiplex PCR protocols were used to screen the presence or absence of hlyA, actA, prfA, and inlA virulence-associated genes among obtained L. monocytogenes isolates. The primers used for amplifications were selected from a previous research (Di Ciccio et al., 2012) and their sequences are listed in Table 1.
The PCR assay was established utilizing standard strain as a positive control (ATCC 19155) and negative control (PCR mixture without DNA template). Initial denaturation was achieved at 94 °C for 3 min followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 2 min and extension at 72°C for 1 min 30 s. Then, the final extension was processed at 72 °C for 15 min (Di Ciccio et al., 2012).
The PCR products were separated by electrophoresis on 1.5% agarose gel (Applichem, Germany, GmbH). The gel was photographed by a gel documentation framework (Alpha Innotech, Biometra) and the data were analyzed by computer software.
Statistical data analysis
The obtained results were analyzed utilizing the descriptive analysis of the Pearson Correlation Test. Significance was recorded at P< 0.05 (Bryman and Cramer, 2002)
RESULTS
In total, 45 (22.5%) Listeria species were isolated from the 200 samples tested in our study when platted onto Oxford agar. Colonies are small (1.0 mm in diameter), grayish, encircled with a black zone.
On ALOA medium, the listeria isolates exhibited bluish-green colonies. Then, pathogenic Listeria spp can be differentiated from other Listeria spp through creating of phosphatidylinositol-specific phospholipase C (PI-PLC), that is demonstrated by the formation of opaque halos around the colonies.
In the present study, minced meat and tilapia fish showed the highest and lowest significant prevalence rates (P< 0.05) among the obtained samples with percentages 56 and 6%, respectively (Table 2).
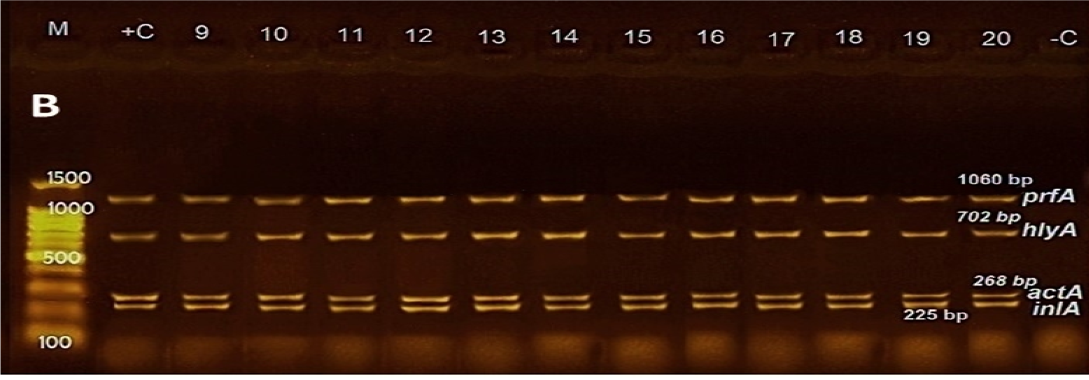
Figure 1: Agarose gel electrophoresis of DNA fragments generated by multiplex PCR with 20 L. monocytogenes isolates. M: DNA molecular weight marker (100 bp); Lanes 1-9: L. monocytogenes strains from minced meat; Lanes 10-14: L. monocytogenes strains from poultry meat, Lanes 15-17: L. monocytogenes strains from fish; Lanes 18-20: L. monocytogenes strains from raw milk; Lanes –C: Negative control; Lanes +C: Positive control.
Table 1: Primers used for PCR reactions
| Target gene | Primer name | Sequence (5=،3) | Amplicon size (bp) |
| hlyA |
HlyAF HlyAR |
CCT AAG ACG CCA ATC GAA AAG CGC TTG CAA CTG CTC |
702 |
| actA |
ActAF ActAR |
GAC GAA AAT CCC GAA GTG AA CTA GCG AAG GTG CTG TTT CC |
268 |
| prfA |
PrfAF PrfAR |
CTG TTG GAG CTC TTC TTG GTG AAGCAA TCG AGC AAC CTC GGA ACC ATA TAC TAACTC |
1060 |
| inlA |
InlAF InlAR |
CCT AGC AGG TCT AAC CGC AC TCG CTA ATT TGG TTA TGC CC |
255 |
Table 2: Detection of Listeria monocytogenes in different food samples
| Food item | Examined samples no. | Listeria spp. positive (%) | No. (%) of identified L. monocytogenes isolates | |
| Rural | Urban | |||
| Minced meat | 50 |
28 (56%)a |
2 (4%) b |
7 (14%)a |
| Poultry meat | 50 |
9 (18%) b |
3 (6%) a |
2 (4%) b |
| Tilapia fish | 50 |
3 (6%) d |
1 (2%) c |
2 (4%) b |
| Raw milk | 50 |
5 (10%)c |
2 (4%)b |
1 (2%)b |
| Total | 200 | 45 (22.5%) | 8 (4%) | 12 (6%) |
Groups within the same column carrying different superscript are significantly different at (P< 0.05).
The dissemination of L. monocytogenes isolates among different types of samples revealed that 9 (18%) of the recovered isolates were from minced meat, 5 (10%) from poultry meat and 3 (6%) from each tilapia fish and raw milk. Totally, the overall prevalence of L. monocytogenes among investigated samples was 10%. The correlation test verified that there was a significant association between the isolation rate for L. monocytogenes and the sample type (P<0.05). Moreover, most isolates were obtained from urban areas with a prevalence rate of 6%.
Regarding multiplex PCR results, All 20 isolates of L. monocytogenes tested for the presence/absence of examined virulence genes harbored all genes and gave their characteristic bands as shown in Figure 1.
DISCUSSION
Listeriosis has been remarked to be one of the emergent zoonotic diseases in recent years, being signed basically through the utilization of contaminated foods and food products. Expanding proofs propose that fundamental aspects of human listeriosis cases are owing to the foodborne transmission of L. monocytogenes (Molla et al., 2004). It remains to be of a great concern to the food manufacturing and regulating agencies worldwide, to the researchers in different authorities, and to the customers of ready to eat foods. Sporadic instances of listeriosis still to appear in different countries and there have been numerous outbreaks of the disease related to food in Egypt (Kamar et al., 2014).
In this assessment, 45 (22.5%) listeria isolates were recovered on ALOA medium from the 200 samples tested giving bluish-green colonies because of the prevalence of a chromogenic compound X-glucosidase, that recognizes b-glucosidase existing in all Listeria spp. This ratio was relatively higher than previous studies in Brazil (16.7%), Iran (20.3%), and Ghana (12.2%) (Monteiro et al., 2013; Haj Hosseini et al., 2014; Kwarteng and Wuni, 2018).
This difference in the obtained data might be because of the divergence in food type understudy, analyzed specimens number and geographical location (Nayakv et al., 2015).
Out of 50 minced meat specimens inspected, more than half (28/50) were contaminated with Listeria species. It may be attributed to the subjection of beef meat samples to insignificant hygienic conditions, throughout slaughtering, evisceration, processing, and vending. This data is in concurrence with the prior reports previously mentioned (Molla et al., 2004; Gebretsadik et al., 2011).
The second source for Listeria spp in our study was the poultry meat with a prevalence rate of 18%; five isolates (10%) were identified as L. monocytogenes. This is nearly by investigations directed in Greece (Sakaridis et al., 2011) and Iran (Zeinali et al., 2017) where L. monocytogenes were isolated from poultry meat with percentages of 38% and 18%, respectively representing contamination evidence linking live production and/or processing environments.
Regarding milk samples, the prevalence of Listeria species differs frequently among various survey reports and has been ascribed to factors such as geographic (ecological) conditions, farm size and management practices (Claeys et al., 2013). In our study, the frequency of Listeria spp. in raw milk was 10%, while in a previous report carried out in Ethiopia (Seyoum et al., 2015) a predominance of over 28% of Listeria spp was detailed. Prior studies mentioned that L. monocytogenes prevalence rates can vary from 0% in raw cow milk (Huehn et al., 2010) to more than 45% (De Reu et al., 2004; El Marnissi et al., 2013).
All three tilapia fish isolates obtained in this study were identified as L. monocytogenes. The previous literatures demonstrated relatively high incidence rates of L. monocytogenes in seafood sample; 7.7% in Iran (Momtaz and Yadollahi, 2013) and 7% in Egypt (Edris et al., 2014). There are two sources of fish contamination with Listeria species, that incorporates; the attack of listeria from intestinal contents to other fish tissues like muscles (Ertacs and cSeker, 2005), and cross-contamination (fish manipulation, using contaminated equipment and inappropriate transport) (Gudbjörnsdóttir et al., 2004).
Inclusively, the isolation rate of L. monocytogenes differed in various regions, giving an overall incidence of 10% (20/200). Moreover, the urban area showed the highest prevalence (6%), even though (Kersting et al., 2010; Amajoud et al., 2018) demonstrated that L. monocytogenes were isolated and overrepresented in high percentages from rural environments. The possible explanation for this discrepancy could be due to the difference in packing methods, handling and geographical difference (Seyoum et al., 2015; Wu et al., 2015).
Concerning PCR results, all genes playing a key role in the pathogenesis of L. monocytogenes (prfA, hlyA, actA, and inlA) were detected in all examined isolates as shown in Figure 1. These data are in concurrence with preceding data conducted in Italy, where prfA and actA genes were predominant (100%) among L. monocytogenes isolates, followed by hlyA (93%) and inlA (60%) genes (Coroneo et al., 2016) illustrating that the pathogenicity island genes LIPI-1, and the inl genes, are invariable parts of the genome of L. monocytogenes (Doumith et al., 2004; Gelbíčová et al., 2015). Moreover, the presence of the hlyA gene in all isolates encodes pore-forming listeriolysin O toxin which necessary for bacterial escape from the phagosomes of host cells into the cytosol (Camejo et al., 2011).
Henriques et al. (1998) detected hlyA genes and actA from L. monocytogenes isolates from all Portuguese ready-to-eat meat samples, which is similar to the findings of this study. Listeriolysin O and actA are associated with the bacterium’s capability of passing the intestinal barrier, cell to cell spread and motility, cell invasion, and intracellular parasitism (Liu et al., 2007b; Rawool et al., 2007).
The high frequencies of virulence genes in the bacterial strains of our study enable adhesion, invasion, and epithelial damage to the human digestive system (Momtaz and Yadollahi, 2013).
Moreover, these results indicate that these isolates possess the properties of virulent strain and their sequences may be further investigated to explore the differences between pathogenic and less pathogenic strains (Abdellrazeq et al., 2014). Thus, the presence of these genes proposes the incidence of a critical public health risk correlated with the consumption of examined food samples sold in Sharkia Governorate contaminated with L. monocytogenes.
On another hand, previous research carried out in Nigeria demonstrated that only 3 genes (hly A, iap, and prf A) among five screened virulence-associated genes were detected in (9/36) L. monocytogenes isolates (Usman et al., 2016).
The prevalence of L. monocytogenes in the minced meat, poultry meat, tilapia fish, and raw milk samples cannot be ignored and we ought not to disregard the danger of listeriosis, because of the consuming of these foods. Moreover, simultaneous recognition of multiple virulence genes in a single assay is desired as it will be helpful for enormous scale investigations aimed at the detection of pathogenic listeria strains.
The authors declare that no conflict of interest exists.
ACKNOWLEDGEMENTS
This work was supported by Animal Health Research Institute, Zagazig, Egypt and Animal Health Research Institute, Dokki, Egypt.
AUTHORS CONTRIBUTION
All authors contributed equally in this work.





