Advances in Animal and Veterinary Sciences
Research Article
Sex Identification Based on Molecular Marker of Chdz and Chdw Gene in Some Species of Indonesian Birds
Yudit Oktanella
Universitas Brawijaya, Indonesia.
Abstract | Birds usually have monomorphic properties, especially the young ones that are difficult to identify their sex based on morphological characteristic. The male birds have two identical sex chromosomes (ZZ), whereas females are heterogametic (ZW), the development of a DNA-based sexing method order widely investigated in several species. In this work, two orders were examined for sex determination, Passeriformes and Psittaciformes. Genomic DNA was isolated from the thirteen feather samples. The amplification of the CHD gene was performed with P2 and P8 sets of universal primers for DNA sexing. The result of this study showed that the target DNA bands were formed in all samples, ranging from 365 bp to 500 bp by DNA electrophoresis using agarose 2%. The single band indicated male and the double bands indicated female with varying DNA fragments thickness. The results of current study indicated both P2 and P8 primers provided reliable results, although few individuals showed the less prominent band.
Keywords | Sex determination, CHD W, CHD Z genes, Passeriformes, Psittaciformes
Received | May 21, 2019; Accepted | July 25, 2019; Published | September 15, 2019
*Correspondence | Yudit Oktanella, Universitas Brawijaya, Indonesia; Email: yuditoktanella@gmail.com
Citation | Oktanella Y (2019). Sex Identification Based on Molecular Marker of CHDZ and CHDW Gene in Some Species of Indonesian Birds. Adv. Anim. Vet. Sci. 7(10): 844-847.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.10.844.847
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Oktanella. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Indonesia is the fourth largest of bird’s biodiversity country after Columbia, Brazil and Peru, with 1,598 species which spread of through the territory (Sukmantoro et al., 2007). Total of 372 of these species (23.28%) are endemic and 149% of them (9.32%) are migratory birds. Breeding in captivity is an effort to meet the needs of population. In bird breeding, sex determination is important but it is relatively difficult through physical observation because more than 50% of birds are monomorphic. So, there are no significant differences in morphology between female and male. In another case, some species of young birds indicate mono-morphologically between male and female, but becoming dimorphic after reach a certain age (Ari, 2015). Sex determination can be examined by surgery, determination of hormone levels, blood plasma protein profile, chromosomal examination, karyotyping, DNA analysis as well as morphological observation (Archawaranon, 2004). However, this method needs 14- 15 day to be analyzed.
The male birds have two identical sex chromosomes (ZZ), whereas females are heterogametic (ZW), several methods have been described using specific primers to amplify homologous sections of two conserved chromo-helicase-DNA-binding (CHD) genes, located on sex chromosomes of all birds (Griffiths et al., 1996; Wang et al., 2007; Bantock et al., 2008). CHD genes have an important role in chromatin remodeling in the control of transcription elongation (Simic, et al., 2003). CHD contains at least two introns which difference in length in the Z and W chromosomes. This allows a significant difference between the target band products from the Z and W chromosomes on DNA electrophoresis agarose (Dubiec & Zagalska-Neubauer, 2006). CHD genes identified by P2-P8 primers designed by Griffits et al. (1998) using Polymerase Chain Reaction (PCR) technique.
Bird sexing is needed for studies of behavior, ecology, evolutionary biology, genetics, and conservation of endangered species (Ito et al., 2003; Wang et al., 2007). This study aimed to determine the sex of various types of birds both chirping and ornamental birds in Indonesia using P2-P8 primers.
Table 1: List of species and type of materials collected for isolation of DNA
| No | No / sample code | Order | Species | Common Name | Indonesian name |
| 1 | 01-02 | Passeriformes | Zosteros flavius | Javan white-eye | Pleci Jawa |
| 2 | 03-04 | Passeriformes | Mirafra javanica | Horsefield Bushlank | Branjangan |
| 3 | 07-10 | Passeriformes | Mirafra javanica | Horsefield Bushlank | Branjangan |
| 4 | 05-06 | Psittaciformes | Agapornis sp | Love birds | Love birds |
| 5 | 11-12 | Psittaciformes | Nymphicus hollandicus | Falk/Cockatiel |
Kakaktua |
MATERIAL AND METHOD
This research is an observational study of several birds species which tested for sex determination in laboratory of Animal Disease Diagnostic, Faculty of Veterinary Medicine, Brawijaya University in the period of September-December 2018th.
Materials and Equipments
Birds feather samples (Table 1), Geneaid extraction kit (Qiagen) Agarose lite (Thermo Scientific). TBE 10x (Biorad), gel Red nucleid acid (Biorad), Gotag green PCR mix reaction (Promega, Cat. No. M7122). 1 kb DNA ladder (Invitrogen), 100 bp DNA Ladder (Invitrogen, Cat. No. 15628019), and sex of primers (P2 dan P8) and disposable items (tubes and tips, thin 200 µL wall tube).
Equipment used for this research were : Centrifuge (Hermle), Horizontal gel electrophoresis (Biorad®), Genecycler / Thermal Cycler (Biorad®), Incubator (Memmert), gel documentation (Biorad®), include scissors, 1.5 mL microtube, 0.1 - 2.5 µL micropipette, 2 - 20 µL, and 20 - 100 µL, white tip, yellow tip, vortex machine, 1.5 mL tube, centrifugation, freezer, incubator, DNA gel electrophoresis (Biorad®), Thermal Cycler (Biorad®).
Methods
Preparation of bird feather samples: Twelve of bird feather was collected from 4 species. 1 mL of plumage was cut into small pieces then put on the 1,5 ml microtube. It was then added with 200 µL lysis buffer, and 20 µL proteinase K and then vortexed for approximately five min and then was incubated at 60⁰C for an hour and the process of vortex and incubation was repeated for 3 times, then it was continued with overnight incubation in 60⁰C.
DNA Isolation: DNA isolation was conducted based on product protocol with slight modification. Elution buffer added into the sample was 50 µL. DNA yielded from this work, then was measured on A260 and A280.
DNA Amplification using Polymerase Chain Reaction (PCR) Methods: DNA sample then amplified using 10 pmol of P2 and P8 primers which indicate CHD genes, The primers used, were shown in Table 2. Amplification process was conducted by this stage, were:
Predenaturation at 940C, 2 min
Denaturationat 940C, 30 sec
Annealing at 580C (gradient ± 4 0C), 30 sec
Extention at 72 0C, 30 sec.
Post extention at 72 0C, 7 min.
Table 2: Primers used in this work
| No | Primers name | Oligonucleotide sequence |
| 1. |
P2 |
5’ TCT GCA TCG CTA AAT CCT TT - 3’ |
| 2 |
P8 |
5’ CTC CCA AGG ATG AGR AAY TG - 3’ |
PCR products obtained from this stage then were visualized on 2% agarose gel electrophoresis with gel red nucleic acid (Biorad®). The 2% agarose gel was transferred to the UV-tray and inserted into the Bio-Rad UV-trans illuminator Gel Doc (Biorad®) to identify the DNA fragments. Then it was analyzed.
RESULT AND DISCUSSION
The DNA produced from isolation process is quantitatively measured on A260 nm and A 280 nm to ensure that the amplification process is supported by an adequate and good quality of DNA. Table 3 showed that the DNA product were sufficient for PCR process.
Table 3: DNA Concentration and Purity
| Sample Code | Species | DNA concentration (ng/ul) | DNA Purity (260/280) |
| 01 | Zosteros flavius | 2.10 | 1.13 |
| 02 | Zosteros flavius | 3.32 | 1.67 |
| 03 | Mirafra javanica | 5.02 | 1.48 |
| 04 | Mirafra javanica | 1.94 | 1.02 |
| 05 | Agapornis sp | 0.41 | 0.91 |
| 06 |
Agapornis sp |
5.35 | 1.74 |
| 07 | Mirafra javanica | 1.13 | 1.08 |
| 08 | Mirafra javanica | - | - |
| 09 | Mirafra javanica | 4.52 | 1.22 |
| 10 | Mirafra javanica | 3.80 | 1.15 |
| 12 | Nymphicus hollandicus | 7.53 | 1.68 |
| 13 | Nymphicus hollandicus | 6.05 |
1.63 |
Table 4: Fragment length of Chromo Helicase DNA-binding (CHD) genes in all samples
| Sample code | Species | Number of band | Fragment l | Decision | |
| 1 | Zosterops flavus | 1 | 478 | Male | |
| 2 | Zosterops flavus | 1 | 495 | Male | |
| 3 | Mirafra javanica | 2 | 500 and 430 | Female | |
| 4 | Mirafra javanica | 2 | 500 and 430 | Female | |
| 5 | Agapornis sp | 1 | 482 | Male | |
| 6 |
Agapornis sp |
1 | 470 | Male | |
| 7 | Mirafra javanica | 2 | 480 and 420 | Female | |
| 8 | Mirafra javanica | 1 | 410 | Male | |
| 9 | Mirafra javanica | 1 | 460 | Male | |
| 10 | Mirafra javanica | 1 | 360 | Female | |
| 12 | Nymphicus hollandicus | 2 | 422 and 360 | Female | |
| 13 | Nymphicus hollandicus | 1 | 400 | Male |
The most common method of acquiring genetic material for molecular sexing is whole-blood sample (Harvey et al., 2006). Unfortunately, blood sampling was avoided in this study in order to minimize the stress during sample collection. Application of genomic extraction from feathers could be a non-invasive method, less time-consuming, and considered as simpler method than the whole-blood sample.
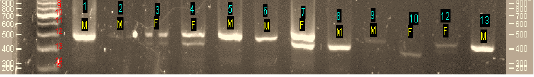
Figure 1: The result of DNA amplification using primer set P2 and P8 (Agarose 2%). Annotation: M = male, F=Female, 1-13: No. of sample
Figure 1 showed that DNA bands were founded in all samples with a variety of target bands, ranging from 360 bp to 500 bp. Single band indicated male and double bands indicated female with varying DNA fragments thickness. Upper bands ranging from 460bp – 500 bp were indicated as male and the lower bands ranging from 360 bp – 430 bp were indicated as the female fragment. It could also be displayed as a single fragment, or double fragments. In this work at code sample of 10, the single band was determined as female because it showed the lower band (360 bp) that resulted on agarose 2% electrophoresis.
Birds have heterogametic female with Z and W sex chromosomes. Single band in males avian was identified as CHD-Z gene (located in Z chromosome). However, the double bands in the females were identified as CHD-Z gene and CHD-W (located in W chromosome). The marker gene in Aves. The CHD gene is on the Z and W chromosomes, which consist of CHD-Z (located on the Z chromosome) and CHD-W (located on the W chromosome) (Dubiec and Zagalska-Neubauer, 2006), distinguished by two sexes (ZZ and ZW). The structure of proteins from CHD-Z and CHD-W are known to have very little differences. Few genes have been found on the W chromosome to identify sex in Aves, but the most commonly used gene is the CHD gene. The key to developing a universal DNA test for avian sexing is to detect the W-chromosome-specific (female-specific) sequence. Nowadays, the mostly-known gene of the avian sex chromosome is The CHD gene. The first gene on the W chromosome, was discovered in 1995 and has been used for sex identification in a wide range of species (Bantock et al., 2008). Full CHD sequences have been reported for only two species, the domestic chicken and Zebra Finch (Griffiths and Korn 1997; Agate et al., 2003), and these genes located only on the W or Z chromosome have not to be identified yet.
Many primers have been designed to recognize different introns in the CHD gene. However, the primers that are mostly used to determine the sex in aves were P2 and P8. This primers have been able to distinguish the sexes of 27 species from 25 families of Struthionidae, Phasianidae, Anatidae, Alcedinidae, Meropidae, Psittacidae, Apodidae, Strigidae, Columbidae, Otididae, Burhinidae, Laridae, Alcidae, Acciptridae, Falconidae, Maluridae, Ardalotidae, Sylvidae, Corvidae, Callaetidae, Paridae, Passeridae and Mure (Uria aagle). The result from this study suggested that CHD-W and CHD-Z genes amplified by P8 and P2 are quite similar in target band, 430bp, and 490bp respectively. In current study, PCR method with the P8 and P2 primers successfully determined the sex from all the tested birds. In spite of successfully sex determination using P2 and P8 primers in this study, the availability of a reliable genetic technique using another recommended primers on many avian will be important for future studies.
Conclusion
P2 and P8 primers encoded of CHD genes were successfully determine the bird sex from 12 samples of local Indonesian birds.
Conflict of interest
The authors declare that there is no conflict of interest in this research article.
References





