Advances in Animal and Veterinary Sciences
Research Article
First Nearly Complete Genome Sequence of Porcine Bocavirus Strain from Malaysia
Chee Yien Lee1, Chew Yee Tan1, Yi Jing Tan1, Jeffrey Wai Nung Wong3, Daniel Mohan Jacob1, Jia Xin Lim3, Gunaselvi Ananthan3, Siti Suri Arshad2, Gayathri Thevi Selvarajah1, Peck Toung Ooi1*
1Department of Veterinary Clinical Studies, Faculty of Veterinary Medicine, University Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia; 2Department of Veterinary Pathology and Microbiology, Faculty of Veterinary Medicine, University Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia; 3NHK Bioscience Solutions Sdn. Bhd., 4 Lorong Chekor, 3rd mile off Jalan Kelang Lama, 58000 Kuala Lumpur, Malaysia.
Abstract | This paper reports the first nearly complete genome sequence of a Malaysian Porcine bocavirus (PBoV) detected in Malaysia from a porcine organ sample. In Malaysia, both Human bocavirus (HBoV) and PBoV have been reported, at prevalence of 5.6% and 90.9% respectively. Most recently, in 2018, PBoV emerged as a zoonosis when a human case of PBoV-related respiratory infection is reported in a child with history of contact with porcine secretion. With this evidence of human infection with PBoV, considering close contact between humans and pigs in the swine industry, further study of PBoV genetic characterization and epidemiology is warranted. This study attempts to obtain the complete genome sequence of a Malaysian PBoV isolate detected in a pig farm in the state of Selangor. A nearly complete sequence was obtained through primer walking and touchdown PCR method. Based on phylogenetic analysis for the nearly complete genome, the Malaysian PBoV strain is genetically characterized as PBoV G3B, sharing a close relationship with a reference PBoV strain from U.S.A. with 98% homology. This finding may facilitate further epidemiological studies and diagnostic development of PBoV in Malaysia.
Keywords | Porcine bocavirus, PBoV, Pigs, Malaysia
Received | April 11, 2019; Accepted | July 18, 2019; Published | August 26, 2019
*Correspondence | Peck Toung Ooi, Department of Veterinary Clinical Studies, Faculty of Veterinary Medicine, University Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia; Email: ooi@upm.edu.my
Citation | Lee CY, Tan CY, Tan YJ, Wong JWN, Jacob DM, Lim JX, Ananthan G, Arshad SS, Selvarajah GT, Ooi PT (2019). First nearly complete genome sequence of porcine bocavirus strain from malaysia. Adv. Anim. Vet. Sci. 7(9): 776-781.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.9.776.781
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Ooi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
The family Parvoviridae is currently classified with two subfamilies: Parvovirinae and Densovirinae, infecting vertebrate and non-vertebrate hosts respectively. Parvovirinae is further classified into five genera: Dependovirus, Bocavirus, Erythrovirus, Parvovirus and Amdovirus (Claude and Fauquet, 2004). Among them, Bocaviruses are considered unique due to the presence of a third open reading frame (ORF) between their non-structural and structural coding regions (Manteufel and Truyen, 2008). Bocaviruses have been detected in human (Allander et al., 2007), swine (Liu et al., 2014; Amimo et al., 2016), canine (Decaro et al., 2012), feline (Lau et al., 2012), bovine (Abinanti and Warfield, 1961), rat (Babkin et al., 2013) and wildlife including gorillas (Lau et al., 2017), California sea lions (Li et al., 2011) as well as bats (Lau et al., 2016). Since the discovery of human bocavirus (HBoV) in 2005 (Allander et al., 2005), it has been associated with human respiratory tract gastrointestinal infections (Schildgen et al., 2008; Kapoor, et al., 2009; Schildgen, 2010; Kantola et al., 2011; Khamrin et al., 2012). In Malaysia, HBoV was first detected in 2012, in a 13-month-old boy with pneumonia and underlying asthma (Etemadi et al., 2012). Subsequent study reported a HBoV prevalence of 5.6% among 125 cases of acute respiratory distress (Tg Rogayah et al., 2014). Most recently, in 2018, PBoV had emerged as a zoonosis; causing respiratory infection in a child with history
Table 1: Primers used in PCR for the PBoV target
| Primer | Orientation | Sequence | Bases | Tm (°C) | Position based on KF025386 (Approx.) |
| a | Forward | GTGCGCCGAGGACTTTGA | 18 | 59.4 | 1951 -> 1968 |
| b | Reverse | CGCAAAGTTACCGCCCAC | 18 | 58.5 | 3245 <- 3262 |
| c | Forward | TATCGACTCTTTGCAGTCTGAC | 22 | 56.9 | 3217 -> 3238 |
| d | Reverse | TGACGTAGATGGTTCCCGGA | 20 | 59.3 | 4801 <- 4820 |
| e | Forward | TTGGGCTGTCTGATCAGGA | 19 | 56.9 | 234 -> 252 |
| f | Reverse | CTGAGATAGTGAGTGATCGGCT | 22 | 58.1 | 1018 <- 1039 |
| g | Forward | ACAGGCAGCCGATCACTCACTAT | 23 | 62.6 | 1008 -> 1030 |
| h | Reverse | CTCGTTCCTCCCATCAGACACTT | 23 | 60.9 | 1728 <- 1706 |
| i | Forward | GGAGATAATGATAGACACGCCTT | 23 | 56.9 | 3 -> 25 |
| j | Forward | GATTTTGTGCTCTACGCTCAAGGAC | 25 | 61.4 | 2060 -> 2084 |
| k | Reverse | CCSRSGAAGAYGTTCCATGCTC | 22 | 62.3 | 2467 <- 2488 |
| m | Reverse | ATAAAGTCTTGATCACCTT | 19 | 47.3 |
3201 <- 3219 |
of contact with porcine secretion (Safamanesh et al., 2018). With this evidence of human infection with PBoV, considering close contact between humans and pigs in the swine industry, further study of PBoV genetic characterization and epidemiology is warranted.
Porcine bocavirus was first identified in lymph nodes of pigs in Sweden, co-detected with Porcine circovirus 2 (PCV2) (Blomström et al., 2009). Detection was reported in wide range of samples including saliva, sera, nasopharyngeal swab, feces, lymph nodes, tonsil, lung, kidney, spleen and liver (Choi et al., 2014; Zhou et al., 2014; Gunn et al., 2015), suggestive of systemic infections. It was linked to gastrointestinal and respiratory signs as well as encephalomyelitis (Pfankuche et al., 2016). The pathogenic pathways of PBoV are rather obscured, with multiple reports of co-infections (Blomström et al., 2009; Shan et al., 2011; Ndze et al., 2013). It was suggested to be indirectly related to porcine post-weaning multisystemic wasting syndrome (PMWS) and other swine diseases as a triggering factor (Blomström et al., 2009). In Malaysia, PBoV was first detected in 2017 in clinically compromised pigs (Jacob et al., 2018). As our continual effort in genetic characterization of Malaysian PBoV isolate, this study attempts to obtain the complete genome sequence by primer walking. In summary, here we described the nearly complete genome of one PBoV field strain isolated from Selangor, Malaysia; and its phylogenetic relationship compared to reference bocavirus strains.
MATERIALS AND METHODS
Sample, DNA Extraction and PCR Assay
From the tissue samples that were detected positive of PBoV previously (Jacob et al., 2018), total DNA was isolated using Patho Gene-spin DNA/RNA Extraction Kit (iNtRON Biotechnology, South Korea). A series of specific PCR primers were designed (synthesized by Bioneer, South Korea) to obtain the initial target sequence to be used as a template to ‘walk’ the length of the target (Table 1). The primers were paired in various combinations resulting in fragments of varying sizes (Table 2). Real-time PCR was performed using NexQ-9300 NEXpro™ qPCR 2x MasterMix Solution (EvaGreen, U.S.A.) with touchdown cycling conditions to increase annealing specificity and yield of the target fragments (Table 3).
Table 2: Combination of PCR primers and their expected PCR product size
| Primer Combination | Nucleotide Positions | Expected Size (bp) |
| i+f | 4->1039 | 1036 |
| e+h | 234->1728 | 1495 |
| g+b | 1008->3262 | 2255 |
| j+m | 2060->3219 | 1160 |
| c+d | 3217->4820 | 1604 |
| a+k | 1951->2488 | 538 |
Table 3: Touchdown qPCR cycling conditions
| Phase | Step | Temperature | Duration | Cycle (s) |
| Denaturation | Melting | 95°C | 300 secs | 1 |
| Touchdown | Melting | 95°C | 15 secs | 16 |
| Annealing |
67°C - 0.5°C/cycle |
55 secs | ||
| Extension | 73°C | 140 secs | ||
| Normal | Melting | 95°C | 15 secs | 19 |
| Annealing | 61°C | 55 secs | ||
| Extension | 73°C | 140 secs | ||
| Extension | Final Extension | 72°C | 600 secs |
1 |
Sequencing and Phylogenetic Analysis
Due to multiple signals being generated from direct sequencing of the fragments, TA cloning using pLUG-Prime TA-Cloning Kit (iNtRON Biotechnology, South Korea) was employed to obtain clean sequences and ascertain whether multiple variants of the PBoV target were present. The near complete genome of PBoV was deduced from the best overlaps spanning the entire sequence of the target PBoV. Phylogenetic analysis of the nucleotide sequences of complete or near complete genome of PBoV and other bocaviruses available in NCBI GenBank database was conducted using MEGA7.0.26 software (Kumar, Stecher and Tamura, 2018) using Maximum Likelihood method based on Tamura-Nei model with 1000 bootstrap replicates (Saitou and Nei, 1987).
RESULTS
A previous selected positive PBoV sample (Jacob et al., 2018) was subjected to real-time PCR and a nearly complete genome sequence comprising 4836 nucleotide with G+C content of 52% was obtained. Sequence fragments generated from PCR of the primer combinations (Table 2) are shown in Figure 1. Nucleotide BLAST analysis of the initial sequencing data indicated that the target had a high homology of 98% with PBoV Group 3 lineage. The near complete genome of Malaysian PBoV had been submitted to GenBank under accession number MK467456. The nucleotide sequence of the near complete genome is shown in Figure 2.
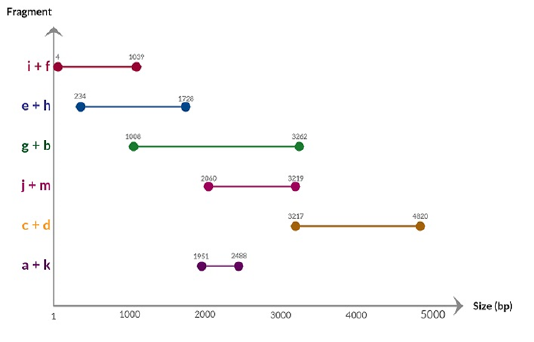
Figure 1: Chart showing the sequence fragments generated from PCR of primer combinations and their respective nucleotide position in the PBoV genome.
The result of phylogenetic analysis comparing Malaysian PBoV isolate with complete or near complete genome for PBoV and other bocaviruses is shown in Figure 3. The Malaysian PBoV isolate MK467456 (indicated by round bullet) formed a clade with PBoV strains KF025382 and KF025386 (indicated by triangle bullets), both isolated from domestic pigs in U.S.A. The result indicates a strong relationship between Malaysian PBoV with these two aboard strains.
DISCUSSION
Grouping system of PBoV varies due to the vast genetic diversities in PBoV isolates. International Committee on Taxanomy of Viruses (ICTV) classified Porcine bocaviruses into four species namely Ungulate bocaparvovirus 1 to 4 (Cotmore et al., 2013). The current ICTV criterion for classification of bocaviruses defines separate species as a group of individuals with less than 95% homologous non-structural gene DNA sequence (Cotmore et al., 2014). Based on that, the NS1 genes of PBoVs would divide them into ten species; but their NP1 genes only divide them into nine (Zhai et al., 2015). Recently, a VP1-based classification has been proposed (Yang et al., 2012), dividing PBoVs into three groups: group 1 or PBoV1, G1 (represented by PBo-likeV), group 2 or PBoV2, G2 (PBoV1/2-CHN and PBoV2-A6), and group 3 or PBoV3, G3 (PBoV3/4-UK, PBoV3/4-HK, and PBoV3C).
From the Figure 3, the Malaysian PBoV isolate MK467456 (indicated by round bullet) showed a strong relationship with the both United States PBoV strains KF025382 and KF025386 (indicated by triangle bullets). This relationship could be linked to the importation source of Malaysian breeding herd, given that majority of the breeder pigs used in Malaysian swine farms are imported from U.S.A (Singh and Fong, 2014). Considering the possibility of introduction of this PBoV G3 isolate through commercial trading by importation of semen and breeder stock, this virus could be expected to be circulating in Malaysian commercial swine herd, as shown by a pilot molecular detection of PBoV in Malaysia reporting a very high prevalence of 90.9% (Jacob et al., 2018). Therefore, understanding the genetic characteristics of the Malaysian isolate of PBoV could contribute to the management of bocavirus infection in the local scenario. Screening of breeder animals and semen for bocavirus prior importation and strict biosecurity measures upon arrival may help to prevent any further introduction of new bocavirus strains into the country. In terms of NS1 gene sequence, these 3 strains share 98% of their nucleotide identity. Thus, this Malaysian PBoV isolate can be classified as same species with reference strains KF025382 and KF025386 as Porcine bocavirus G3B.
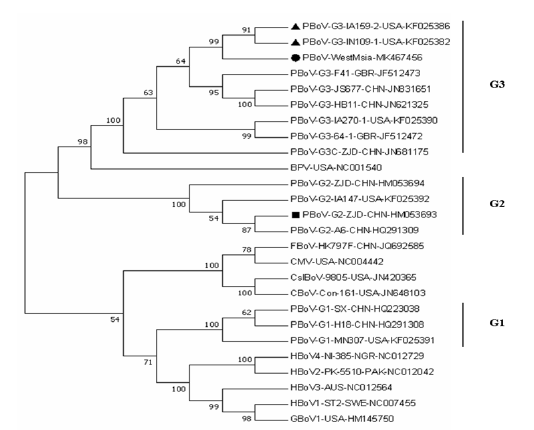
Figure 3: Phylogenetic tree of Malaysian PBoV isolate MK467456 with 25 nucleotide sequences of complete or near complete genome of PBoV and other bocaviruses. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The vertical lines indicate PBoV grouping based on VP-1 classification. Round bullet designates Malaysian PBoV sequence generated from the present study; triangle bullets designate the two U.S.A. PBoV strains
Bocaviruses from G2 and G3 are more distantly related to the HBoV, as compared to G1 which shares a large clade with HBoV. Although the Malaysian PBoV is only distinctly related to HBoV, possible zoonotic risk still cannot be ruled out. Furthermore, strain HM053693 (indicated by square bullet) reported to be closest to the strain in the first human case of PBoV (Safamanesh et al., 2018), is classified as PBoV G2 that shares a clade with G3. This might suggest potential zoonotic risk. Thus, this genetic characterization of Malaysian PBoV G3B isolate could be useful for the start of epidemiology studies of PBoV in Malaysia: further studies to determine whether PBoV G1 and G3 is present in Malaysia, as well as whether the Malaysian PBoV strain poses zoonotic risk.
CONCLUSION
Publication of near complete genome sequences allows researchers to trace the presence of new, important emerging viruses and determine their genetic distribution to plan relevant control programs. With the characterization of Malaysian Porcine bocavirus isolate as strain PBoV G3B having close relationship with U.S.A. reference strains KF025382 and KF025386, we suggest that routine screening of imported breeders could be helpful to monitor the status of new emerging disease in Malaysian swine farms. Further studies are required to establish the clinical impact of the current Malaysian PBoV G3B strain as well as to determine presence of other PBoV strains in Malaysia. In conclusion, PBoV is present in Malaysia and the Malaysian isolate is confirmed to be of PBoV G3B strain.
STATEMENT OF ETHICAL STANDARDS
This research project was approved by the Institutional Animal Care and Use Committee (IACUC), with reference number UPM/IACUC/AUP–R063/2014.
ACKNOWLEDGEMENT
The authors are thankful to the Government of Malaysia and its Ministry of Higher Education for funding this research through the Fundamental Research Grants Scheme (FRGS) (Grant no. FRGS 2014-1-5524541) and Universiti Putra Malaysia through the Putra Grant – Putra Graduate Initiative (Grant no. GP-IPS/2016/9492500).
Conflict of interest
The authors declared that no conflict of interest in this research study.
authors contribution
LCY, TCY, TYJ contributed in manuscript writing and data interpretation. LJX, DM and JWWN conducted the research work. OPT, SS, GTS and DM contributed to methodology design and manuscript editing.
REFERENCES






