Advances in Animal and Veterinary Sciences
Research Article
Mycoplasma wenyonii: A Causative Agent of New Mastitis in Dairy Cows
Salam Abd-Esmaeel, Basima Abdulfatah Albadrani*
College of Veterinary Medicine, University of Mosul, Mosul-Iraq.
Abstract | This is the first reported molecular detection of hemoplasmas (Mycoplasma wenyonii) DNA in cow’s milk samples. Mycoplasma wenyonii caused a recurrent and persistent udder infection which failed to respond to treatment as Escherichia coli or Streptococcus mastitis on many farms from different regions in Nineveh province of Iraq. The study recorded about 66% PCR confirmed cases over the last year that suggests it was undiagnosed earlier. M.wenyonii should be considered in a differential diagnosis in any cow presenting with mastitis, particularly if there has been a recent reduction in milk yield. All milk samples infected with M.wenyonii organisms identified on the basis of the 16S rRNA gene sequence (deposited in GenBank under accession number MK335902, MK335904). The DNA sequence had 100% identity to previously reported M. wenyonii16S rRNA gene in blood samples of the same dairy cows.
Keywords | Mycoplasma wenyonii, Mastitis, PCR, Milk, Dairy cows.
Received | February 10, 2019; Accepted | March 14, 2019; Published | April 24, 2019
*Correspondence | Basima Abdulfatah Albadrani, College of Veterinary Medicine, University of Mosul, Mosul-Iraq; Email: basima1971@yahoo.com
Citation | Abd-Esmaeel S, Albadrani BA (2019). Mycoplasma wenyonii:a causative agent of new mastitis in dairy cows. Adv. Anim. Vet. Sci. 7(6): 480-483.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.6.480.483
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Abd-Esmaeel and Albadrani. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Hemotrophic Mycoplasmas (hemoplasmas) are uncultivable eperythrocytic emerging pathogens that are found in a wide range of domestic and wild animals (Fujihara et al., 2011). M.wenyonii is a hemotrophic mycoplasma that causes acute and chronic infections in cattle (Santos et al., 2012). Signs of dysgalactia were identified in sows infected with M.suis beginning 1 day after parturition and lasted 4 to 6 days, this resulted in a mean piglet preweaning mortality rate of 18% because of starvation (Strait et al., 2012). In recent years, the number of reports describing a new type of udder infection could be going widely undiagnosed on-farm and could be to blame for recurrent mastitis cases, a combination of clinical signs in cattle comprising pyrexia, hind limb and/or udder edema, pre-femoral and supra-mammary lymphadenopathy with decreased milk production consistently associated with M.wenyonii infection (Strugnell and McAuliffe, 2012; Gladden et al., 2016; Basima and Baraa, 2016). M.wenyonii is a contagious pathogen with a vector, the main route of spread is thought to be via needles or arthropods and events involving blood contact are suspected to be involved in transmission. Due to lack of in vitro cultivation systems for the isolation of these haemoplasmas has hampered and delayed their detection and characterization in humans and animals (Rani et al., 2018). Little is understood about the molecular mechanisms of pathogenicity of M.wenyonii. To deal with this problem, we used PCR methods for the direct detection of M.wenyonii DNA from cow’s milk samples.
MATERIALS AND METHODS
One hundred dairy cows (ages > 3years) from six farms of different regions in Nineveh province of Iraq were included in the study for the period from September 2017 to September 2018. Case history and clinical signs of udderinfection were recorded for cows failed to respond to treatment as E.coli or Streptococcus spp. Mastitis. Milk samples (10 ml) were collected from each infected dairy cow after properdis infection of teats and removing first drops; milk sample was placed in plain tube and stored at -20°C prior to DNA extraction. DNA was purified from milk samples. A total Genomic DNA was extracted from 10 ml of each milk sample with commercially available kit according to the manufacturer’s recommendations (Bioingentech veterinary extraction, purification kit. Chille), and stores -20°C until further analysis. The presence of amplifiable DNA was confirmed for each sample using a 16S rRNA gene. Initially, a conventional PCR assay was carried out using specific designated primers (Veterinary Laboratory Agency (AHVLA), UK), for the 16S rRNA gene of bovine hemoplasmas - Forward primer:, 5’-ATATTCCTACGGGAAGCAGC-3’, equivalent to nucleotide numbers 328 to 347 of M. wenyonii- Reverse primer: 5’-ACCGCAGCTGCTGGCACATA-3’, equivalent to nucleotide numbers 503 to 522 of M. wenyonii amplified a 195 bp and 173 bp product for M. wenyonii and ‘Candidatus M. haemobos’, respectively (Nishizawa, et al., 2010) with few modification, 2 µl of DNA sample from milk, 1 µl Forward primer (10 Pico mole) and 1 µl Reverse primer (10 Pico mole), 4µl of 5× Taq master mix, 12 µl PCR Grade water in a 20 µl final PCR reaction. The thermo cycling program for PCR used initial denaturation at 94°c for 30 sec., annealing at 57°c for 30 sec., extension at 72°C for 1min and final extension 72°c for 2 min., cooling 4°c. All amplicons (PCR products) were run in an electrophoresis chamber on a 1.5% agarose gel in TBE buffer and visualized under UV light. In a second step, Amplified fragments were randomly selected for sequence determination using forward and reverse PCR primers. The obtained sequences were compared for similarity with sequences in the GenBank database using the Basic Local Alignment Search Tool (BLAST) program (National Center for Biotechnology Information, National Institute of Health). Denaturing gradient gel electrophoresis (DGGE) of a 16S ribosomal DNA was used to separate PCR products from Milk DNA according to differences in primary sequence (Macrogen, Inc., South Korea public biotechnology company). New sequences were submitted to GenBank.
Statistical Analysis
Calculations were performed with the SPSS software Analyse-it (Analyse-it Software Ltd., Leeds,United Kingdom) for Microsoft Excel.
RESULTS
According to clinical observations, udder infection usually starts in dairy cows with significant edema on one quarter and can occur at any point in lactation. Occasionally more than one quarter can be involved. Within 2 days of this edema being identified, clots will usually then appear in the milk. The swelling is different to that caused by trauma or E.coli, as Mycoplasma wenyonii will result in a spongy ’pitting type edema’ around the quarter which will form a ‘pit’ or dent when pressed with a finger. This dent will then take about five-10 seconds to disappear (Figure 1).
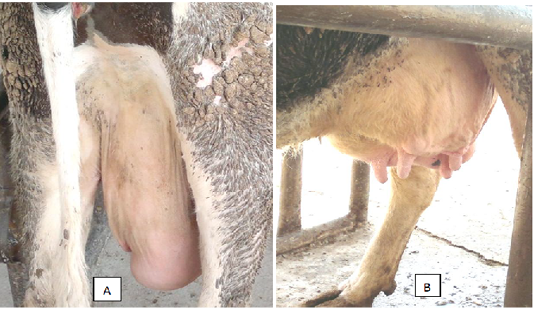
Figure 1: A cow with pitting type edema around the quarter (A) and swollen teats (B) was infected by Mycoplasma wenyonii
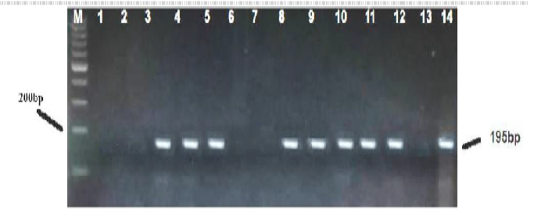
Figure 2: PCR based detection of M. wenyonii 16S rRNA in dairy cows. The amplified 195 bp product, from the positive sample control (line 14) and positive test samples (lines 3,4,5,8,9,10,11,12) were subjected to electrophoresis in 1.5%-agarose gel and stained with ethidium bromide. Marker 200 (M) and a negative control free of DNA template (line 13) were run in parallel.
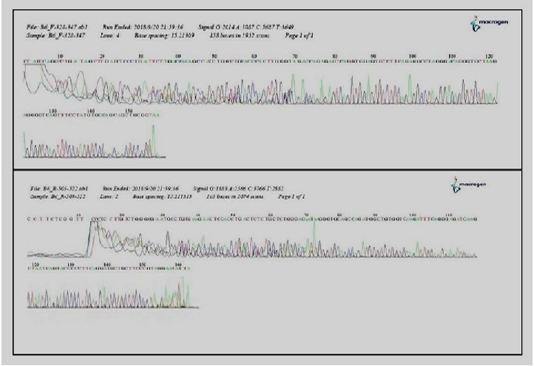
Figure 3: Detection and sequencing of Mycoplasma wenyonii agents in cattle milk by (DGGE) of a 16S ribosomal DNA PCR.
Sixty six milk samples (66%) were found positive for specific PCR. Results of amplification showed that bands on the Agarose gel appeared at the concentration of 1.5% and with expected result reaction of 195 base pairs (Figure 2). The result of the sequencing of the products isolated by DGGE evidenced the presence of M.wenyonii in eight samples. The results of DGGE for two of the DNAs tested were shown (Figure 3). All milk samples infected with M.wenyonii organisms identified on the basis of the 16S rRNA gene sequence (deposited in GenBank under accession number MK335902, MK335904). This means M.wenyonii could be the underlying cause of culls for mastitis.
DISCUSSION
Mastitis is an age-old problem and even today it continues to be the most significant disease of dairy cattle. M.wenyonii, could be the underlying cause of recurrent and persistent udder changes. It’s an enigmatic and incompletely understood organism, and there is a growing body of evidence suggesting that it may contribute to the clinical signs described in this article, particularly in dairy cows. With advances in research and improvements in the interpretation of assay results, PCR and other molecular biology techniques are likely to gain a more prominent place in mastitis diagnostics in the future. Various PCR assays were used to detect difficult cultivatepathogenslike Mycobacterium avium ssp. Paratuberculosis (Johne’s disease) (Over et al., 2011), Mycobacterium bovis (Sreevatsan et al., 2000; Antognoli et al., 2001), Brucella spp. (Romero and Lopez-Goñi 1999; Sreevatsan et al., 2000), Coxiellaburnetii (Muramatsu et al., 1997), Cryptosporodium spp. (Laberge and Griffiths, 1996), and Mycoplasma spp. (Baird et al., 1999). The diagnosis was confirmed on 100 milk samples by the use of polymerase chain technology using a pair of primers designed based on Nishizawa et al. (2010). It is well established that the haemoplasmas attach to the surface of the red cells but may under certain conditions penetrate this host cell (Messick, 2004). M.wenyonii is a different type of Mycoplasma spp. to the more commonly recognised Mycoplasma bovis and presents differently. Many mycoplasmal pathogens exhibit filamentous or flask-shaped appearances and display prominent and specialized polar tiporganelles that mediate attachment to host target cells (Hoelzle, 2008). These are the complex structures, composed of a network of interactive proteins, designated adhesions, and adherence accessory proteins. These proteins cooperate structurally and functionally to mobilize and concentrate adhesions at the tip and permit mycoplasmal colonization of mucous membranes and eukaryotic cell surfaces (Rani et al., 2018). M.wenyonii causes an infection of the lymph system in the udder and subsequently reduces milk expression. That, in turn, leads to raised somatic cell counts and clots in the milk, the majority of somatic cells in milk are leukocytes, the concentration of leukocytes in milk increases with increased immune response to mastitis caused by pathogenic bacteria (Balsom, 2017). PCR can be a useful diagnostic aid when combined with other currently available tools and herd data.The advantages of PCR method for detection of M.wenyonii DNA in milk samples are a rapid and specific test, the preserved samples can be used, samples are not needed to be collected separately, also samples from groups of cows can be pooled to help cut down on costs. Identifying sources of mastitis in a single cow or bulk tank sample, identifying sources of mastitis in cows already being treated, finding sub-clinical cases before they become clinical or chronic. PCR products of approximately 195 bp were obtained for all M.wenyonii isolates tested. The use of denaturing gradient gel electrophoresis (DGGE) of a 16S ribosomal DNA to separate PCR products according to differences in primary sequence enabled differentiation of all M.wenyonii isolates analyzed. The DNA sequence had 100% identity to previously reported M. wenyonii 16S rRNA gene (deposited in GenBank under accession number MK335902, MK335904) in blood samples of dairy cows (Esmaeel and Albadrani, 2019). This molecular technique on milk samples is the first study conducted on haemotropic Mycoplsma spp. which can be a new causative agent of mastitis in dairy cows.
ACKNOWLEDGEMENTS
This work was kindly supported by Dean of Veterinary Medicine College, We are grateful to the Genomic Core Facility at University.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHORS CONTRIBUTION
All authors contributed equally.
REFERENCES





