Advances in Animal and Veterinary Sciences
Research Article
Molecular-Genetic Analysis of Cows Genetic Structure and Determination of Genealogical Relatedness Level of Bulls of Modern Dairy Breeds
Volodymyr Ladyka1*, Yuriy Skliarenko1, Yulia Pavlenko1, Olena Metlytska2, Irina Ivankova2
1Sumy National Agrarian University, 160, H. Kondratiiev St., Sumy, Ukraine; 2Institute of Animal Breeding and Genetics nd. a. M.V.Zubets of NAAS, 1, Pogrebnyak P.L St., Chubinske village, Boryspil district, Kyiv region, Ukraine
Abstract | As a result of molecular-genetic analysis of two cow populations – Ukrainian Brown Dairy breed (UBD) and Sumy interbreed type of Ukrainian Black-and-White breed (SITUBWD) in the technique of ISSR analysis with the use of the fragment of microsatellite loci with trinucleotide core motif (AGC)6 C (S1) as an amplification primer, there were identified 23 DNA-patterns. It was determined that genetic locus with the identified size of 1350 bp was monomorphic in animals of SITUBWD occurring in all study samples, however, in UBD breed cows the amplicons of this size were not detected at all (p<0.001). The population-genetic analysis of the two studied animal groups (taking into account maternal origin) by molecular-genetic testing in the ISSR-PCR technique revealed essential statistically significant differences. The total average number of PCR products obtained from the cows of the newly developed Sumy interbreed type of Ukrainian Black-and-White breed was 15,050 being 12,450 in Ukrainian Brown Dairy breed (p <0,001). A rather high level of genetic similarity between the animals of Ukrainian Brown Dairy and Sumy interbreed type of Ukrainian Black-and-White Dairy (0,768) is quite possibly connected with the influence of the unique aboriginal Lebedynsky breed on which maternal base they were developed.
Keywords | Cow population, Molecular-genetic analysis, Genetic structure, Ukrainian Black-and-White breed
Received | October 08, 2018; Accepted | February 18, 2019; Published | March 25, 2019
*Correspondence | Volodymyr Ladyka, 1Sumy National Agrarian University, 160, H. Kondratiiev St., Sumy, Ukraine; Email: v.i.ladyka@ukr.net
Citation | Ladyka V, Skliarenko Y, Pavlenko Y, Metlytska O, Ivankova I (2019). Molecular-genetic analysis of cows genetic structure and determination of genealogical relatedness level of bulls of modern dairy breeds. Adv. Anim. Vet. Sci. 7(5): 405-411.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.5.405.411
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Ladyka et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
The purposeful breeding of the brown dairy breed is based on the theory and practice of animal breeding supplemented with the modern elements of large-scale and individual selection in the form of new system for evaluating the genotype of Lebedynsky breed with the improving breeds and on the openness of the population to better heredity of the brown cattle. By combining in the process of selective breeding the animals with high milk yields and optimal milk composition and the elements of the dairy type, the conditions for a significant competitiveness of the modern brown cattle population were created (Baschenko et al., 2017; Ladyka et al., 2012).
During the last 10 years the Holstein breed cows have been keeping a notable advantage as for the average milk
yields. A three-fold decline of pedigree cows of Ukrainian Brown Dairy breed (from 1 395 to 479 heads) and the use of Brown Swiss bulls only provided the increase of their productivity (by 55.8%). However, the localization of a small (less than 250 cows) active (pedigree) part of Ukrainian Brown Dairy population just in two breeding herds does not provide valid grounds for asserting its leadership in productivity and prospects for interbreed genetic improvement among other indigenous dairy breeds (Bondarchuk, 2018).
The absence of sires of Ukrainian Brown Dairy significantly complicates the selective breeding work, making it practically impossible. It is the use of bulls of the related Brown Swiss breed that enables to preserve brown cattle herd in the region (Buchanan and Lenstra, 2015).
As a result of the long selective breeding work, Ukrainian Black-and-White Dairy breed was developed, including five interbreed types. The development of Sumy interbreed type of Ukrainian Black-and-White Dairy breed was accompanied by the use of several black-and-white breeds: Ukrainian Black-and-White Dairy and Holstein, based on Lebedynsky breed (Ladyka et al., 2012; Novikov et al., 2016).
Unfortunately, the prospects of Sumy interbreed type development are not clear which coincides with the general tendency as for Ukrainian Black-and-White Dairy. At SE Sumy State Breeding Center there are no sires of Ukrainian Black-and-White Dairy breed. So the breeding farms have no possibility to use planned pedigree sires of indigenous selection of Holstein breed (Sklyarenko et al., 2015).
The population condition of Ukrainian Brown Dairy and Sumy interbreed type of Ukrainian Black-and-White Dairy in Sumy region is characterized mainly by the presence of a substantial number of animals with a high percentage of genes (more than 87.5%) of Brown Swiss and Holstein breeds (Mokhnacheva, 2018).
Currently, gene pools of the indigenous cattle breeds as for the peculiarities of the genetic structure based on the genes quantitative characteristics are almost not investigated. Therefore, the identification of genes and their mutations determining the direction and degree of quantitative trait loci (QTL) is profitable due to the reduction of the generation interval, early introduction of the pedigree stock, selection of parental pairs according to the set of certain genotypes and getting the offspring with the corresponding genetic potential as for the basic productivity indicators (Zinovyeva et al., 2016; Kopylov et al., 2017).
One of the directions of the use of these immunogenetic studies in breeding is the study of the genetic structure of the breeding populations, herds, lines by marker genes. Studies of many scientists proved the reliability of using the blood genetic markers for the cattle genotype evaluation (Iso-Touru et al., 2016).
The purpose of the article is to conduct a molecular genetic analysis of the cows of the above-mentioned breed populations and to determine the level of the genealogical relationships between the bulls of Holstein and Brown Swiss breeds used on the breed livestock farms and the ancestors of the lines.
Materials and methods
To determine the genealogical relatedness of the lines we used the data of the genealogical composition of the bulls populations of Brown Swiss, Ukrainian Brown Dairy and Holstein breeds, which took part in the creation of Ukrainian Brown Dairy breed and Sumy interbreed type of Ukrainian Black-and-White dairy breed.
For the genealogical analysis of the bulls population of Brown Swiss breed the data of the origin of 158 sires belonging to 13 lines were used: Vigata 083352 (11 head), Destini 118612 (11), Distinkshna 159523 (21), Eleganta148551 (42), Eleima 110327 (4), Kontsentrata 106157 (6), Laddi 125640 (6), Lailasana 131528 (4), Oregona 086356 (6), Peivena 136140 (10), Reinera 124788 (1), Stretcha 143612 (30), Hilla 76059 (6).
For the genealogical analysis of the bulls population of Ukrainian Brown Dairy breed we used the data of the origin of 31 sires of the following lines: Destini 118612 (5), Eleganta 148551 (5), Eleima 110327 (2), Lailasana 131528 (7), Oregona 086356 (5), Stretcha 143612 (3), Suprima 124652 (4).
For the genealogical analysis of the bulls population of Holstein breed the data of the origin of 366 sires belonging to 21 lines were used: R.Soveringa 198998 (21), S.T. Rokita 252803 (36), V.B.Aidiala 1013415 (3), М. Chifteina 95679 (20), І.S. Reflekshna 121004 (2), Suprima 503024/288659 (10), Starbarka 352790 (23), Aivengo 1189870 (27), Metta 502096/1392858 (23), P.F.A. Chifa 1427381 (40), Eleveishna 1491007 (51), Valianta 1650414/502383 (28), Hanovera 1629391/502304 (5), Ingansera 343514503283 (1), Montfrecha 91779 (27), P.Astronavta 1458744 (15), Elbrusa 897 (9), Siteishna 1492073/267150 (11), Kutlasa 340909 (3), Kavalera 1620273 (4), Bella 1667366 (7).
For the genetic characterization of cows sample of Ukrainian Brown Dairy breed and Sumy interbreed type of Ukrainian Black-and-White Dairy breed, we collected 1 ml of blood from the jugular vein into the disposal conical plastic tubes with 3.8% solution of sodium citrate.
The cows were selected from the herd of the State enterprise “Research Farm of the Institute of North East Agriculture of NAAS” in the amount of 20 head of Ukrainian Brown Dairy breed and Sumy interbreed type of Ukrainian Black-and-White dairy breed. The animals of Ukrainian Brown Dairy breed came from 10 sires of four Brown Swiss breed lines: Eleganta 148551 (Abel АТ 593920645, Poliden US 193950, Balero АТ 225588461, Yakob АТ 317506961); Distinkshna 159523 (Evgen АТ 720448311, Erlander 722113211, Basik US 197201, Beni СН 120059571329), Peivena 136140 (Rubal АТ 357552666), Laddi 125640 (Sonet UA 136). The cows of Sumy interbreed type of Ukrainian Black-and-White dairy breed came from 15 sires of five Holstein breed lines: Eleveishna 1491007 (Frits US 135326066, Pilgrim US 136932888, Chipotl US 51870141, Eigg US 135556243), Chifa 1427381 (Dzhesko DE 346539791, Leros АТ 909528547, Georg US 132950253, Dzhupiter DE 27640964506), Valianta 1650414/502383 (Zhelanny UA 509), Siteishna 1492073/267150 (Berkut UA 5254), Starbaka 352790 (Manat 909534347, Dzhokus 113080315, Tikon US 136135816, Shyrli NL 447860719, Manat АТ 909534347).
DNA was extracted from the animal blood after obtaining leukocyte sediment from leukocyte sedimentation using 5% Chelex-100 @ (Sota, 2005; Walsh et al., 1991).
DNA amplification with primer ISSR was performed using the commercial kit “Fermentas” (Vilnius, Lithuania). The reaction mixture composition was: reaction buffer – 2.5 μl; 100 пM primer - (0,5-1,0) μl; from 2 to 4 activity units of Taq-polymerase – (0.1-0.2) μl; (1-2 ng) DNA sample – (1-3) μl; deionized water (to bring the volume of the mixture to 25μl). Not more than 0.5-1 ng of DNA sample (stock solution of DNA in the ratio of 1:10) was used. The primer synthesis used in the inter microsatellite analysis technique (ISSR) was performed by Fermentas (Vilnius, Lithuania) under order.
The nucleotide structure and temperature condition of the primer was: S1 3’-AGCAGCAGCAGCAGCAGCC-5’, acting as a forward and reverse initiator of the amplification in PCR.
The amplification was carried out in the thermocycler «Tertsik» («DNA-technology», Moscow, Russia). The amplification program with the primer S1 was: 1 cycle: 94°C - 4 minutes; 2 - 31 cycles: 57°C - 2 minutes; 72°C - 4 min; 94°C - 1 minute; 32 cycles: 57°C - 3 min; 72°C - 7 minutes.
Electrophoretic separation of amplified DNA regions by the ISSR technique was performed in 2% agarose gel in a single Tris Borate Electrophoresis buffer, according to the methodological recommendations (Yurchenko et al., 2018; Maniatis et al., 1984). Gels were coloured with 0.5% solution of bromide ethidium for 10 minutes. The visualization of the electrophoregram was performed on the transilluminator in the ultraviolet spectrum at the wavelength of 340 nm. The gel was photographed by Canon camera using the orange filter. The control of the size of amplification products on the gel electrophoregrams was performed with the use of the molecular weight marker 1 kb -Ledderplus (“Fermentas”, Vilnius, Lithuania). Only those PCR products were analyzed, which were clearly reproduced on the gels in the range of molecular weights relative to the marker: from 200 bp up to 3000 bp in the course of 3 repetitive amplification reactions. The received profiles were processed in the standard computer program GenAlex6 (Buys, 1990; Peakall and Smouse, 2006). The construction of the cluster was carried out according to the values of genetic distances in the MEGA 4 program (Tamura et al., 2004).
The determination of the genetic relationship of the lines was carried out according to Push-Shaporuzh conventional method. The principle of the method is to calculate the genealogical lines with a common ancestor, starting from the first line – sire and dam. The classification defines the inbreeding type according to the ancestor line with the common ancestor (Zinovyeva et al., 2016; Kopylov et al., 2017).
Results and Discussion
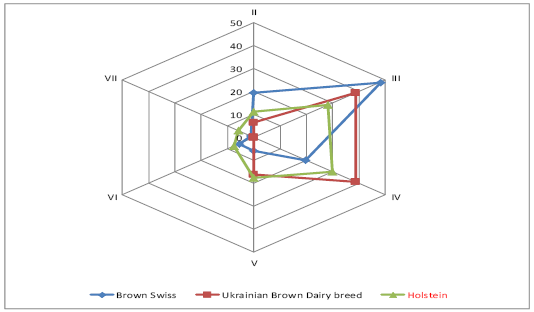
The genealogical structure of Ukraine’s brown cattle was formed under the influence of the intensive use of Brown Swiss sires during the last 35 years, therefore the names of the lines of Ukrainian Brown Dairy breed are identical to Brown Swiss ones. The relation between the lines depends on the number of the sires and their remoteness from the ancestor. The majority of the bulls used for the development of Ukrainian Brown Dairy breed and Sumy interbreed type of Ukrainian Black-and-White Dairy breed was in lines III-IV to the ancestor (Figure 1). It should be noted that most of Brown Swiss bull sires are in line III to the ancestor.
Most Brown Swiss bulls of Vigata 083352 (82%) and Elegantna 148551 (26%) lines are in line II to the ancestor (Figure 2). The bulls of Distinkshna 159523 (57%), Elegantna 148551 (62%), Stretcha 143612 (70%) lines are in line III to the ancestor. 27% of bulls of Stretcha143612 line are in line III to the ancestor, 67% of bulls of Laddi 125640 are in line V to the ancestor, 50% of bulls of Kontsentrata 106157, 100% bulls of Oregona 086356, 50% bulls of Hilla 76059 are in line VI to the ancestor. There are bulls of Kontsentrata 10615 and Hilla 76059 (40%) only in line VII to the ancestor.
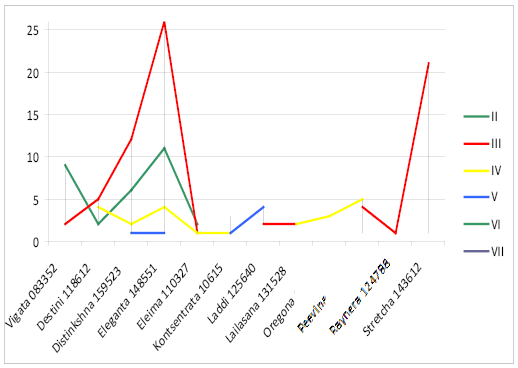
In line II to the ancestor there are only bulls of Ukrainian Brown Dairy breed of Eleganta 148551 (40%) (Figure 3).
Bulls of Eleganta 148551 (60%), Suprima 124652 (50%), Stretcha 143612 (67%), Eleima 110327 (100%) are in line III to the ancestor. In line IV to the ancestor there are bulls of Oregona 086356 (100%), Suprima 124652 (50%), Lailasana 131528 (57%). 80% of bulls of Destini 118612 are in line V to the ancestor.
The majority of animals of Ukrainian Brown Dairy breed were in lines III-IV to the ancestor, indicating the possibility of consolidating desired economic-beneficial traits of offspring due to the effective selection of parents.
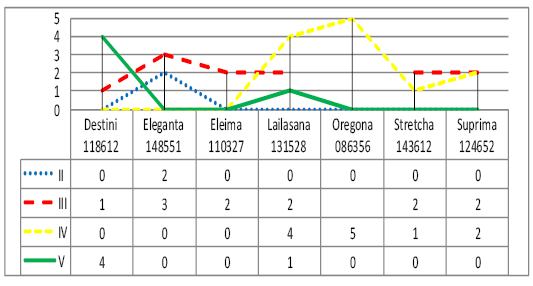
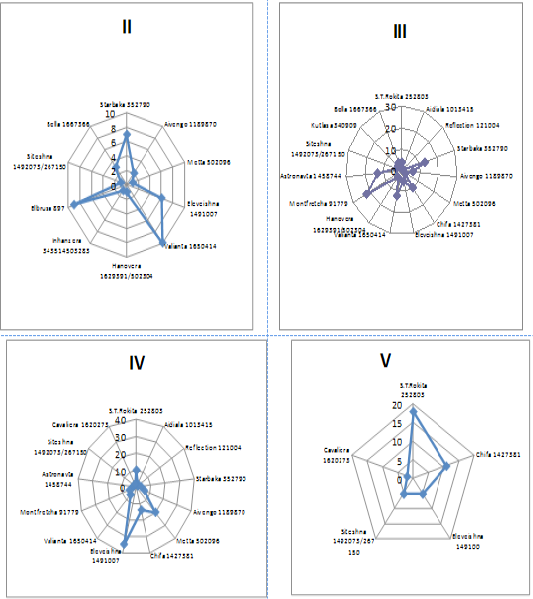
The genealogical structure of bulls used for the development of Sumy interbreed type of Ukrainian Black-and-White Dairy breed was formed under the influence of the intensive use of Holstein bull sires during the last 30 years, therefore the names of the lines are identical to Holstein ones. According to the research results the bulls used in the livestock of Sumy interbreed type of Ukrainian Black-and-White Dairy breed belonged to twenty-one genealogical formations (Figure 4). Most animals were in lines II-III to the ancestor that also indicates the possibility of consolidating desired economic-beneficial traits of offspring under the effective selection of parents. The most numerous were the bulls of six lines – S.T.Rokita 252803, Chifa 1427381 and Eleveishna 1491007.
Large amount of Holstein bull sires are in line II to the ancestor: Valianta 1650414 - 44%, Elbrusa 897 – 100%, Starbaka 352790 – 30%, Eleveishna 1491007 – 12%. The bulls of Starbaka 352790 (57%), Chifa 1427381 (25%), Valianta 1650414 (43%), Montfretcha 91779 (82%), Astronavta 1458744 (87%) are in line III to the ancestor. In line IV to the ancestor there are 28% of bulls of S.T.Rokita 252803, 87% of Metta 502096, 35% of Chifa 1427381, 69 % of Eleveishna 149100. In line V to the ancestor there are 50% of bulls of S.T.Rokita 252803, 44% - Aivengo 1189870, 28% - Chifa 1427381. 100% of bulls of Suprima 563024 are in line VI to the ancestor and 100% of M.Chifteina 95679 bulls are in line VII.
As a result of molecular-genetic analysis of two cow populations – Ukrainian Brown Dairy breed (UBD) and Sumy interbreed type of Ukrainian Black-and-White breed (SITUBWD) in the technique of ISSR analysis with the use of the fragment of microsatellite loci with trinucleotide core motif (AGC)6 C (S1) as an amplification primer, there were identified 23 DNA-patterns (Table 1), which were clearly reproduced during three repetitive polymerase chain reactions under standard conditions. Sizes of certain DNA fragments with the use of the primer of the above structure fluctuated in significant limits: from 250 bp to 1600 bp.
Table 1: Comparison of frequency and size of DNA fragments in ISSR technology with primer S1 in cows of two populations
| № DNA fragment | Size of DNA fragment | Cattle populations | |
| Ukrainian Brown Dairy, n=20 | Sumy interbreed type of Ukrainian Black-and-White, n=20 | ||
| 1 | 1600 |
0.0000 |
0.0500 |
| 2 | 1500 |
0.0000 |
0.0500 |
| 3 | 1350 | 0.0000 |
1.0000*** |
| 4 | 1230 | 0.1000 |
0.3000 |
| 5 | 1100 | 0.1000 |
1.0000*** |
| 6 | 1050 | 0.0000 | 0.0000 |
| 7 | 1000 |
0.3000* |
0.0000 |
| 8 | 960 | 1.0000 | 0.9000 |
| 9 | 900 | 0.7000 |
1.0000* |
| 10 | 850 | 0.0000 | 0.0000 |
| 11 | 830 |
0.8000 |
0.5000* |
| 12 | 800 | 0.9000 | 1.0000 |
| 13 | 770 |
0.3000* |
0.0000 |
| 14 | 700 | 0.7000 |
1.0000* |
| 15 | 670 | 1.0000 | 1.0000 |
| 16 | 650 |
0.9000 |
1.0000 |
| 17 | 620 | 0.8000 | 1.0000 |
| 18 | 580 | 0.0000 | 0.1000 |
| 19 | 560 | 0.3000 | 0.4500 |
| 20 | 530 | 0.9500 | 1.0000 |
| 21 | 475 | 0.8000 | 0.9000 |
| 22 | 450 | 0.8500 | 1.0000 |
| 23 | 350 |
0.2500** |
0.0000 |
| 24 | 325 | 0.7500 | 0.8000 |
| 25 | 250 | 0.9500 |
1.0000 |
Note: the difference is statistically significant upon Fisher’s criterion:
* -p<0,05; **-p<0,01; ***-p<0,001
The most statistically significant differences were found for the animals of two compared groups at frequencies of amplicons of large molecular size in the range of 700-1600 bp. DNA-fragments with the size of 1500 and 1600 bp occurred with frequencies that didn’t exceed the value of 5% in animals of Sumy interbreed type of Ukrainian Black-and-White breed being absent in cows of Ukrainian Brown Dairy breed. Genetic locus with the identified size of 1350bp was monomorphic in animals of SITUBWD, occurring in all study samples, however, in UBD cows the amplicons of this size were not detected at all (p<0.001).
The amplification product with the size of 1100 bp was characteristic of all SITUBWD representatives while its frequency in animals of Ukrainian Brown Dairy was 0,1 (p<0.001). Amplicons with the sizes of 770 bp and 1000 bp occurred with the frequency of 0,300 in cows of UBD and were not found in the studied SITUBWD animal sampling.
Unessential but statistically significant difference in the distribution of frequencies of the detected DNA fragments (p<0,05) between the animals of the two groups was determined by amplification products sized 700, 830, 900 bp respectively. It should be noted that DNA-patterns with the size of 580 bp occurred with the frequency of 0,100 in SITUBWD cows and were absent in UBD animals. Low molecular mass DNA fragment of 350 bp occurred in almost 25% of investigated UBD breed species and wasn’t found in the representatives of SITUBWD (p<0,01).
Using the basic parameters of the population-genetic analysis of the two studied animal groups by molecular-genetic testing in the ISSR-PCR technique we revealed essential statistically significant differences (Table 2). Thus, the total average number of PCR products obtained from the cows of the newly developed Sumy interbreed type of Ukrainian Black-and-White breed was 15,050 while it was 12,450 in Ukrainian Brown Dairy breed (p <0,001). The index of within-group similarity has high informative value for animals of artificial populations. For the genetic marker with the dominant type of display used in the research it is defined by the number of coincidental amplicons of the total number of identified DNA fragments. In a certain way, this indicator reflects the level of genetic consolidation of the studied group of animals and their genetic similarity among themselves. The level of within-group similarity for cows of Ukrainian Brown Dairy breed was 0.778, while the cows of Sumy interbreed type of Ukrainian Black-and-White Dairy breed had a similarity index at the level of 0.914 (statistically significant difference, p <0.001). The fact of low basic indices of the genetic diversity of SITUBWD population according to the number of calculated parameters attracts attention. Animals of the newly developed type showed a low value of the theoretically calculated heterozygosity, which was only 0.100, the same indicator for UBD cows being - 0.360 (p <0.001). The total number of identified genetic loci in SITUBWD cows (13,678) was higher than that of Ukrainian Brown Dairy breed (9,151), but the proportion of polymorphic loci in animals of the newly developed type was almost three times smaller than that of the other population cows (p <0.001).
According to the overwhelming number of studies in the field of population genetics of various biological objects, especially of agricultural purposes, one of the important elements of selection is to maintain population heterozy gosity at an optimal level. By numerical value, this level may differ significantly from the selected species as well as from the class of the selected genetic marker.
Table 2: Main genetic-population characteristics of the two cow populations by the results of ISSR-S1 polylocus analysis
|
Breeds (N) |
Genetic-population records | ||||
| Average number of amplicons | Level of group similarities | Heterozygosity | Number of genetic loci | Share of polymorphic loci | |
|
UBD (20) |
12,450***±0,531 |
0.778*** |
0,360а |
9,151 |
0,782 |
|
SITUBWD (20) |
15,050а±0,266 |
0.914а |
0,100*** |
13,678 | 0,196 |
Note: а the difference is likely to be relative to the indicator indicated by the superscript if there are several parameters compared
***-p<0,001
In our case, the characteristic of 9 to 13 polymorphic genetic loci can be compared with the results of immunogenetic testing of agricultural populations (Figure 5).
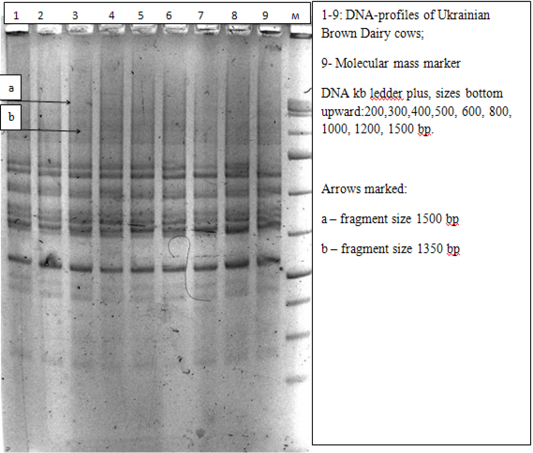
Figure 5: Electrophoregram of DNA amplification products of animals of Ukrainian Brown Dairy breed obtained with ISSR-S1 primer in 2% agarose gel
Thus, the value of heterozygosity of animals of Sumy interbreed type of Ukrainian Black-and-White Dairy breed is precariously low (0,100), which is undoubtedly related to the effect of the “bottle’s neck”, the influence of the genotype of line founders on the newly developed group of animals. It is well known that the main task of the herd selective breeding is to support the optimal level of heterozygosity of the population. Both excessive increase and rapid decrease of heterozygosity have unfavourable consequences hindering the normal herd functioning and affecting the main productivity indicators, especially reproducible animal qualities. Selection contributes to the moderate inbreeding regime in the population. From the point of view of breeding, genetically homogeneous animals represent the greatest value for the breeding business, since the probability of characteristics splitting in the offspring of the next generations from such animals is kept to the minimum. A rather high level of genetic similarity between the animals of Ukrainian Brown Dairy and Sumy interbreed type of Ukrainian Black-and-White Dairy (0,768) is quite possibly connected with the influence of the unique aboriginal Lebedynsky breed, the sires of which made a significant contribution to the formation of their general genetic profile.
Thus, modern breeding and molecular-genetic approaches aimed at creating an optimal structure of each breed, the development of algorithms for the use of new genealogical groups of farm animals for heterozygosity effects, especially in terms of technological adaptability typical for the representatives of aboriginal breeds, should contribute to the preservation of breeds with a limited gene pool, promote their competitiveness at the market.
Conclusions
1. It was revealed that sires of Ukrainian Brown Dairy breed belonged to seven genealogical formations, Brown Swiss - up to thirteen lines, which were in lines III-IV to the ancestor, indicating the possibility of consolidating the desirable economic-beneficial characteristics through clear plans of selecting sires due to the pedigree stock.
2. The intensive use of Holstein bulls for absorptive cross-breeding contributed to the formation of an appropriate linear structure of Ukrainian Black-and-White Dairy breed, which currently includes twenty-one genealogical formations. Most of the animals are in lines II-III to the ancestor that indicates the possibility of consolidating desired economic-beneficial characteristics of the herd when successfully selected.
3. As a result of molecular-genetic analysis of two cow populations - Ukranian Brown Dairy breed (UBD) and Sumy interbreed type of Ukrainian Black-and-White breed (SITUBWD) in the technique of ISSR analysis with the use of the fragment of microsatellite loci with trinucleotide core motif (AGC)6 C (S1) as an amplification primer, there were identified 23 DNA-patterns. It was determined that genetic locus with the identified size of 1350 bp was monomorphic in animals of SITUBWD occurring in all study samples, however, in UBD breed cows the amplicons of this size were not detected at all (p<0.001).
4. The population-genetic analysis of the two studied animal groups (taking into account maternal origin) by molecular-genetic testing in the ISSR-PCR technique revealed essential statistically significant differences. The total average number of PCR products obtained from the cows of the newly developed Sumy interbreed type of Ukrainian Black-and-White breed was 15,050 being 12,450 in Ukrainian Brown Dairy breed (p <0,001).
5. The level of within-group similarity for cows of Ukrainian Brown Dairy breed was 0.778, while the cows of Sumy interbreed type of Ukrainian Black-and-White Dairy breed had a similarity index at the level of 0.914 (statistically significant difference, p <0.001).
6. The proportion of polymorphic loci in SITUBWD cows was three times smaller than that in UBD ones. However, the total number of identified genetic loci in SITUBWD cows (13,678) was higher than that of Ukrainian Brown Dairy breed (9,151).
7. A rather high level of genetic similarity between the animals of Ukrainian Brown Dairy and Sumy interbreed type of Ukrainian Black-and-White Dairy (0,768) is quite possibly connected with the influence of the unique aboriginal Lebedynsky breed on which maternal base they were developed.
Acknowledgments
This article is prepared within the framework of the grant “Justification of the methodology of preserving the brown cattle population in the conditions of the north-eastern region of Ukraine” (No0117U004253).
Conflict of Interest
The authors declare no conflict of interest.
Authors Contribution
Conceptualization – Volodymyr Ladyka, data curation and formal analysis – Yulia Pavlenko and Olena Metlytska, investigation and methodology – Irina Ivankova and Yuriy Skliarenko.
References