Advances in Animal and Veterinary Sciences
Research Article
Gentamicin enhances toxA expression in Pseudomonas aeruginosa isolated form cow mastitis
Atheer Abdul Razzaq1, Ansam Khalid Mahmood1, Nuhad Mohammed Hammed2, Kifah A. Jasim3, Zaid Saifuldeen Abdulqader3, Harith Jabbar Fahad Al-Mathkhury4*
1College of veterinary medicine, University of Baghdad; 2Al-Anbar education directorate, Ministry of Education; 3Epidemiological Unit, Central public health laboratory, Ministry of Health; 4Department of Biology, College of Science, University of Baghdad, Iraq.
Abstract | The present study was undertaken in order to investigate the role of gentamicin in the gene expression of toxA in Pseudomonas aeruginosa isolated from cow mastitis. A total of ten P. aeruginosa strains originally isolated from cows infected with mastitis. Agar dilution methodology was performed to determine the minimal inhibitory concentration of gentamicin, all of which developed resistance toward gentamicin. The findings presented here demonstrated that all these strains harboured toxA depending on PCR-based assay. Nonetheless, RT-PCR technique revealed a wide variation in expression of toxA. Moreover, the cultivation of P. aeruginosa in the presence of gentamicin, significantly (P< 0.05), induced the expression of toxA, in addition to the possibility of enhancing the virulence of this bacterium. In conclusion, using gentamicin to treat infections caused by P. aeruginosa may participate in more severe outcomes.
Keywords | Gentamicin, Mastitis, Pseudomonas aeruginosa, toxA
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | August 15, 2018; Accepted | October 04, 2018; Published | October 17, 2018
*Correspondence | Harith Jabbar Fahad Al-Mathkhury, Department of Biology, College of Science, University of Baghdad, Iraq; Email: harithfahad@scbaghdad.edu.iq
Citation | Razzaq AA, Mahmood AK, Hammed NM, Jasim KA, Abdulqadir ZS, Al-Mathkhury HJF (2018). Gentamicin enhances toxA expression in Pseudomonas aeruginosaisolated form cow mastitis. Adv. Anim. Vet. Sci. 6(12): 526-530.
DOI | http://dx.doi.org/10.17582/journal.aavs/2018/6.12.526.530
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2018 Razzaq et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Pseudomonas aeruginosa is an adaptable bacterial pathogen related with a wide range of infections in humans and animals. Due to the innate capacity of resistance to antimicrobial agents, this bacterium is greatly difficult to treat. What’s more, such resistance is being progressively a problematic issue because of increasingly development of resistance to agents regarded as powerful therapeutic options (Kerr and Snelling, 2009).
This pathogen has a propensity to initiate sudden clinical, or perhaps, subclinical cases of mastitis outbreaks in a number of cows within short period of time. A group of signs and symptoms accompany the acute cases of bovine mastitis with noticeable udder swelling; fever, in addition to abnormal watery as well as clotty milk, which contains flakes or blood. Moreover, severe signs might be seen as well, including toxemia, however, high mortality rate occurred regardless of aggressive treatment. Yet, those being survived will be unfit for productive use (Park et al., 2014).
Locally, in Iraq, twenty-two samples comprising eight milk and 14 udder wounds were positive for P. aeruginosa out of 140 samples (Saleh et al., 2016). Furthermore, Naser and Ismaeel, (2016) alongside with Neamah, (2017) separately reported that P. aeruginosa represented 44% of cow mastitis.
Exotoxin A is the most important virulence factor elaborated by most of the P. aeruginosa strains. It plays a crucial role in the P. aeruginosa pathogenesis. Markedly, this toxin has been recognized as very toxic for mammalian cells (Dong et al., 2015). It has the ability to inhibit protein synthesis of the bacterial cell via the ADP-ribosylation of cellular elongation factor 2. Eventually, this toxin action leads to restricted damage in tissue and therefore bacterial invasion (Michalska and Wolf, 2015). The toxA gene is a chromosome characteristic genetic sequence of P. aeruginosa responsible for regulating exotoxin A synthesis, has been extensively used in molecular detection of P. aeruginosa (Dong et al., 2015).
Subinhibitory concentrations of antibiotics are known to provoke extensive transcriptional changes in bacteria (Ohlsen et al., 1998), to our knowledge, this is the first report that investigating the influence of gentamicin at sub minimal inhibitory (sub MIC) on gene expression of toxA. Therefore, the main goal of this study is investigating the effect of gentamicin at sub MIC on the gene expression of toxA of P. aeruginosa isolated form cow mastitis.
Materials and Methods
A total of ten P. aeruginosa isolates were obtained from microbiology laboratory, College of veterinary medicine, University of Baghdad. Originally, these isolates were isolated from milk samples obtained from mastitis-affected cows. Nevertheless, re-identification was accomplished using the conventional biochemical tests (Gram stain, lactose fermentation, catalase, oxidase, IMViC, motility, and hemolysis pattern) in addition to VITEK 2 compact and API 20NE (bioMe´rieux, France) identification systems.
Determination of Minimum Inhibitory Concentration of Gentamicin
Different concentrations of gentamicin (2 – q1024 µg/ml) were used to estimate the MIC following agar dilution method and the results were interpreted in accordance to the guidelines of Clinical Laboratory Standards Institute (2016), P. aeruginosa ATCC 27853was used as a quality control strain.
Molecular Detection of ToxA
In order to amplify a fragment of toxA (352 bp), the DNA was amplified in a thermal cycler (Bio rad, USA) using the primers (FP 5´-GGTAACCAGCTCAGCCACAT-3´, RP 5´-TGATG TCCAGGTCATGCTTC3´) (Banerjee et al., 2017). Amplicon was visualized in 1.5% agarose gel electrophoresis.
Culture Method
Tryptone soy broth (TSB; HiMedia, India) with or without subinhibitory concentrations of gentamicin were inoculated with each isolate at concentration compatible to MacFarland (1-1.5 × 108 CFU/ml) at 37°C for 24 hr. thereafter, they were sent for RT-PCR study.
Real-Time Quantitative Polymerase Chain Reaction
Extraction of RNA was performed using protocol of TRIzol™ Reagent (Rio et al., 2010). cDNA was created following the manufacturer’s instructions of the RT–PCR kit supplied byInvitrogen.
The gene (toxA) expression in P. aeruginosa culture was accomplished via RT-q PCR technique by Magnetic induction cycler (Bimolecular system, Australia) using primers listed in Table 1. Nonetheless, the reaction mixture is demonstrated in Table 2. Control samples were created by using 10.5 µl nuclease-free water and no cDNA template. However, 16SrRNA was employed to normalized the gene expression of toxA.
Table 1: qPCR Primers used throughout this study (Goldsworthy 2008)
| Gene Primer | Sequence (5´ - 3´) |
| 16srDNA | F- ACCTGGACTGATACTGACACTGA |
| R- GTGGACTACCAGGGTATCTAATCCT | |
| toxA | F- GGAGCGCAACTATCCCACT |
| R- TGGTAGCCGACGAACACATA |
| Reactant | Volume (µl) |
| iQ SYBR Green supermix | 12.5 |
| forward primer | 1 |
| reverse primer | 1 |
| nuclease-free water | 9.5 |
| cDNA template | 1 |
Reaction conditions for 16SrRNA and toxA genes amplification were achieved as described by Goldsworthy (11). For every qPCR a plate read-out was taken after each cycle. Moreover, a melting curve using 1°C increments was also performed following the 40 cycles in order to determine that the correct gene was amplified.
The relative expression ratios were calculated by using the cycle threshold (Ct) of the 16SrRNA as the calibrator (n-fold expression = 2−ΔCt, where ΔCt represents the difference between the Ct of the toxA and the Ct of the 16SrRNA).
Statistical Analysis
Data were presented as the mean of three replicates ± standard error of the mean. The statistical significance of toxA gene expression differences between P. aeruginosa isolates was determined based on repeated analysis of variance (ANOVA) using the statistical program IBMSPSS version 25. P values of <0.05 were regarded as significant (Paulson, 2008).
Results
The identification was confirmed for all isolates included in the present work depending on the results summarized in Table 3. In regard to MIC, all ten isolates were resistant to gentamicin (MIC ≥ 16 µg/ml) in accordance to CLSI breakpoints (Table 4).
Table 3: Results of identification of study isolates.
| Id | Test | Result |
| 1 | Gram stain | Negative |
| 2 | MacConkey agar | Pale colonies |
| 4 | Oxidase | Negative |
| 5 | Catalase | Positive |
| 6 | Indole Test | Negative |
| 7 | Methyl Red Test | Negative |
| 8 | Voges-Proskauer | Positive |
| 9 | Citrate Utilization | Positive |
| 10 | Motility Test | Positive |
| 11 |
β Hemolysis |
Positive |
Table 4: MIC of gentamicin of P. aeruginosa isolates included in present work
| Isolate code | MIC µg/ml |
| P1 | 32 |
| P2 | 64 |
| P3 | 64 |
| P4 | 128 |
| P5 | 32 |
| P6 | 32 |
| P7 | 128 |
| P8 | 32 |
| P9 | 64 |
| P10 | 64 |
The current findings revealed that all isolates of P. aeruginosa harboured toxA (Figure 1). Moreover, these ten isolates developed very low expression levels of mean Ct value of 28.0 ± 1.02.
Pseudomonas aeruginosa 16SrRNA displayed, similarly, moderate levels of expression, giving mean Ct values of 22.93 ± 0.67.
When P. aeruginosa isolates grown in the presence of gentamicin; toxA expression mean has increased significantly (P< 0.05) over their spouses before the addition of gentamicin. Fold change ranged from 5.11 – 323.36 and it varied significantly (P < 0.05) among the isolates as it is depicted in Figure 2.
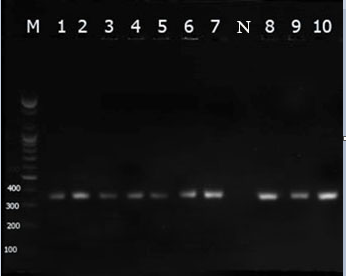
Figure 1: Visualization of toxA gene (352 pb) by 1.5% agarose gel analysis. The shown bands are representative of PCR products amplified from P. aeruginosa (lanes 1 -10), negative control (lane N), lane M represents 100 bp DNA ladder.
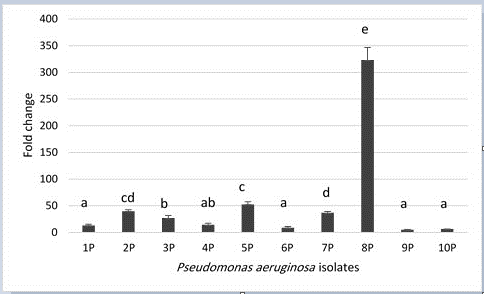
Figure 2: Fold change of toxA gene expression levels in P. aeruginosa isolates from bovine mastitis. P= 4.56 × 10-21, LSD0.05= 13.008. Columns with similar letter have insignificant differences. Error bars represent standard error of mean.
Discussion
Observational data investigated the correlation between virulence and antibiotic resistance has established a simultaneous rising of both of them (Schroeder et al., 2017). However, the current findings are in agreement with a study done by Neamah, (2017) as he stated the toxA was detected in all P. aeruginosa strains that isolated from cow mastitis. While, Raziq, (2017) has detected toxA gene in 84% of the isolates.In compatible with our results, Banerjee et al. (2017) in a study done in South Bengal reported that,nearly, 5.4% of bovine subclinical mastitis were due to toxA containing P. aeruginosa.
Markedly, the link between toxin production and antibiotic resistance has been analyzed by many authors. Ohlsen et al. (1998) demonstrated that some methicillin-resistant S. aureus isolates produced up to 30-fold more alpha-toxin in the presence of 10 mg of methicillin per ml than in its absence. The authors explained such an interaction may induce signal transduction mechanisms, resulting in activation of the hla promoter. However, it cannot be explained by increased levels of the regulatory molecule RNA III. Moreover, Tofik (2011) confirmed that toxin production by S. aureus in the presence of oxacillin was significantly up-regulated. Another hypothesis was adopted by Kimmitt et al. (2000), SOS-inducing antimicrobial agents, particularly the quinolones, trimethoprim, and furazolidone, were shown to induce toxin gene expression.
The regulatory mechanisms govern the interconnection between virulence determinants and genes responsible for the antibiotic resistance is highly complicated. Occasionally these mechanisms are believed of as distinct events.Yet, form the genetic point of view, such regulatory mechanisms are interweaved and connected. Frequently, the expression of antibiotic resistance genes can be influenced by the regulatory mechanisms of virulence determinants and vice versa. Unluckily, the host can affect this gene regulation directly or indirectly via many forms. Of these, stress response, environmental sensing, post-transcriptional modifications, ribo-regulation, multi-networks of regulation, and formation of biofilm govern by quorum sensing (Schroeder et al., 2017).
Conclusion
The findings demonstrated in the present work confirmed that growing P. aeruginosa in the presence of gentamicin induced toxA expression and might enhance the virulence of this pathogen; upon that, including gentamicin in the treatment regimen of infections caused by P. aeruginosa might severely worsen the outcomes.
Acknowledgements
The authors would like to thank the workers in the microbiology laboratory, College of veterinary medicine, University of Baghdad for their kind cooperation in providing the bacterial isolates.
Conflict of interests
No conflict of interest.
Authors contribution
Atheer Abdul Razzaq, Ansam Khalid Mahmood, and Nuhad Mohammed Hammed: Sample collection, and manuscript preparation.
Kifah A.Jasim and Zaid Saifuldeen Abdulqader: Measurements, Acquisition and Analysis of data.
Harith Jabbar Fahad Al-Mathkhury: Study conception and design, Interpretation of data and Drafting of manuscript.
All authors read and approved the final manuscript.
References





