Advances in Animal and Veterinary Sciences
Research Article
Genetic Improvement of Litter Size in Four Goat Breeds in Egypt Using Polymorphism in Bone Morphogenetic Protein 15 Gene
Hanim Shabaan Mohammed Heikal1, Walaa Slouma Hamouda Abd El Naby2*
1Genetics and Genetic Engineering, department of Animal Husbandry and Animal Wealth Development, Faculty of Veterinary Medicine, Sadat City University, Egypt; 2Genetics and Genetic Engineering, department of Animal Husbandry and Animal Wealth Development, Faculty of Veterinary Medicine, Alexandria University, Egypt.
Abstract | Goats are preferred as a source of good quality meat and milk. Their production efficiency can be improved and increased by introducing highly fecundity goats. The aim of this study was to investigate the polymorphism of BMP15 fecundity gene by PCR-RFLP and single nucleotide polymorphism (SNP) in four goat breeds in Egypt (Zaraibi, Baladi, Damascus, and Alpine) and their association with litter size. The genomic DNA was extracted from the whole blood, and 141 base pair fragment encoding BMP15 exon 2 was amplified, purified and sequenced. The sequencing data analysis revealed four SNPs for high litter size does and three SNPs in low litter size. Three SNPs in BMP15 exon 2 at nucleotide position 757 A>C, 762 G>A and 769 G>C in Alpine, Zaraibi, and Baladi respectively. These nucleotide changes associated with amino acid substitution. Furthermore, one novel non-synonymous mutation has been detected in all higher litter size studied goat breeds and lead to change amino acid where Glutamic acid>Glutamine. The SNPs and amino acid substitutions were detected and repeated in higher and lower litter size animals, which can be used as marker-assisted selection for both traits in the four goat breeds under study.
Keywords | Goat, Breeds, SNPs, BMP15 gene, Reproduction
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | July 23, 2017; Accepted | August 22, 2017; Published | October 07, 2017
*Correspondence | Walaa Slouma Hamouda Abd El Naby, Genetics and Genetic Engineering, department of Animal Husbandry and Animal Wealth Development, Faculty of Veterinary Medicine, Alexandria University, Egypt; Email: walaa.slouma@yahoo.com
Citation | Heikal HSM, El Naby WSHA (2017).Genetic improvement of litter size in four goat breeds in egypt using polymorphism in bone morphogenetic protein 15 gene. Adv. Anim. Vet. Sci. 5(10): 410-415.
DOI | http://dx.doi.org/10.17582/journal.aavs/2017/5.10.410.415
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2017 Heikal and El Naby. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Goats are most domestic species that are widely spread all over the world and have an important economic role in developing countries (Adriana et al., 2010). In Egypt, goats are preferred small ruminant used as a source of meat and milk for the farmer. They are distributed in the country, especially in the Nile River Valley and Delta region (Galal et al., 2005). Increasing productivity of goats will contribute to improve the standard of rural people living (Adu et al., 1988).Several authors reported different figures for litter size in different goat breeds (Abdel-Raheem 1998; Campbell, 1994; Song et al., 2006). Hamed et al. (2009) showed that selection would lead to improving goat litter size at birth.
Three fecundity genes were identified in sheep, one of these genes is Bone morphogenetic protein 15 (BMP15) which is a member of the TGF-β superfamily, also known as FecX or GDF9B (Galloway et al., 2000). The BMP15 gene is an ovary-derived growth factor that is essential for follicular development (Hanrahan et al., 2004). This gene plays an important role in female fertility through promoting the growth and maturation of ovary, promoting granulosa cells mitosis, prevention of granulosa cell apoptosis, regulation of the sensitivity of granulosa cells to follicle-stimulating hormone (FSH) action, contributing in determination of the number of eggs that are ovulated, promotion of developmental competence of oocytes and kit ligand expression stimulation (Juengel et al., 2002; McNatty et al., 2003; Moore et al., 2003; Shimasaki et al., 2004; Persani et al., 2014). Moreover, ewes with two inactive copies of the BMP15 gene (homozygous for this mutation) are infertile with primary ovarian failure. While, ewes with a single inactive BMP15 (heterozygous for mutations) are fertile and the effects of these mutations on ovulation rate are additive (Davis et al., 1991; 1993; Hanrahan et al., 2004).
Goat breeds in Egypt, lacking the molecular characterization required for establishing adequate utilization of genetic variation in the development of its production. However, the identification of polymorphism and DNA markers associated with reproductive traits could be used as marker-assisted selection (MAS) which lead to genetic improvement to increase litter size and reproduction efficiency (Ghaffari et al., 2009).
The aim of this study is to screen for BMP15 gene polymorphism by PCR-RFLP and single Nucleotide Polymorphism (SNP) using DNA sequence analysis in four goat breeds in Egypt (Zaraibi, Baladi, Damascus, and Alpine) and their association with litter size.
MATERIALS AND METHODS
Sample Collection and Genomic Dna Extraction
Blood samples were collected from the jugular vein of fifty does from the four breeds (14/ Baladi, 15/ Zaraibi, 11/ Damascuse and 10/Alpine). The Four goat breeds were obtained from Sakha Animal Production Research Station, Kafr El Sheikh governorate, Egypt. Approximately 10 ml of blood was collected and stored at -20o C before further processing. All selected does were healthy with low (less than two kids/litter) or high (two or more kids/litter) litter size. Genomic DNA was extracted using QIAamp DNA Blood Mini Kit (QIAGEN, Germany) according to manufacturer’s instruction.
PCR Amplification of BMP15
The 141 bp fragment encoding BMP15 (exon 2) was amplified using primer 5’- CACTGTCTTCTTGTTACTGTATTTCAATGAGAC-3’ and 5’- GATGCAATACTGCCTGCTTG- 3’ (Kasiriyan et al., 2011). PCR amplification was performed in a total reaction volume of 50 µl which containing 4 µl genomic DNA, 5 µl 10X buffer, 1 µl dNTPs mix (Thermo scientific, Lithuania), 1 µl of each primer (50 nm) (Invitrogen, USA), 0.6 µl of Taq DNA polymerase 500U (Thermo scientific, Lithuania) and 38.4 µl dH2O. The amplification reaction conditions was carried out using 35 cycles at 94°C for 5 min, followed by 45 sec, 62°C for 40 sec, 72°C for 45 sec, followed by 72°C for 10 min. The PCR product of each sample and 100 bp DNA ladder (Thermo scientific, Lithuania) were loaded in 2% agarose gel stained with ethidium bromide. The electrophoresis was carried out and the electrophoresis gel was visualized and photographed using gel documentation system (InGenius, Syngene Bio Imaging, USA).
Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (Pcr-Rflp)
All PCR products of BMP15 gene were digested with HinfI restriction enzyme (Promega, USA). The PCR-RFLP was carried out in reaction volume 20 µl consist of: 10 µl PCR product, 2 µl Restriction Enzyme 10X Buffer, 0.2 Acetylated BSA, 10µg/µl, 7.3 µl dH2O and 0.5 µl of HinfI restriction enzyme 10u/µl. The reaction was incubated at 37°C for 1hr and the products were separated by 3% agarose gel stained with ethidium bromide and visualized on UV transilluminator.
Pcr Purification for Dna Sequencing and Data Analysis
The PCR products were purified and sent for sequencing (3PCR products/each breed) in Invitrogen Company, Germany. The obtained sequences were analyzed using Bio Edit (7.1.3) program to detect Single Nucleotide Polymorphisms (SNPs) from the aligned sequences and their translated amino acid. Sequence alignments were compared with Capra hircus breed Black Bengal BMP15 gene that available in the Gene Bank database (accession number gb|EU888137.1|).
RESULTS
Amplified Fragment of Bmp15 Gene
The PCR amplification BMP15 gene for fifty does which have at least one record for litter size, were screened for BMP15 mutations in the present study. PCR amplification of BMP15 gene yielded a fragment of 141 bp as shown in Figure 1.
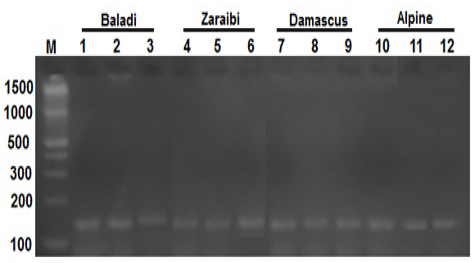
Figure 1: Lane 1-12 represents the PCR product of BMP15 gene (141-bp) of four goat breeds and Lane (M) is a DNA marker
Genotyping of Bmp15 gene Using Rflp Technique
Analysis of HinfI-PCR-RFLP of BMP15 (141-bp) indicate presence of one uncut monomorphic band among all studied animals (higher and lower litter size).
Table 1: Single nucleotide polymorphism and amino acid variations of four goat breeds under study.
| SNPs | Litter size | Amino acid changed in studied Goat breeds | |||
| D | B | A | Z | ||
|
751 G>T and 752 G>C 752 G>C |
Low High |
251 Gly>Cys - |
251 Gly>Ala 251 Gly>Ala |
- - |
- 251 Gly>Ala |
|
757 A>C |
Low High |
- - |
- 252 lys> Gln |
- 252 lys> Gln |
- 252 lys> Gln |
|
762 G>T 760 G>C and 762G>A |
Low High |
- 254 Glu>Gln |
- 254 Glu>Gln |
254 Glu>Asp 254 Glu>Gln |
- 254 Glu>Gln |
|
769 G>A and 769 G>C 769 G>C |
Low High |
257 Glu>Gln - |
257 Glu> lys 257 Glu>Gln |
257 Glu> lys 257 Glu>Gln |
- 257 Glu>Gln |
|
805 G>C
|
In all goat breeds (low and high) |
269 Glu>Gln
|
269 Glu>Gln
|
269 Glu>Gln
|
269 Glu>Gln
|
Glu= Glutamic acid, Gln= Glutamine, Gly= Glycine, Cys= Cysteine, lys=Lysine Asp=Aspartic acid and Ala= Alanine. Z for Zaraibi, B for Baladi, A for Alpine and D for Damascus.
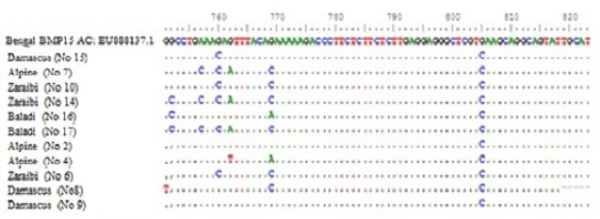
Figure 2: Nucleotide alignment showing SNPs in four goat breeds under study. Does No. (10Z, 6Z, 14Z, 17B, 7A, 15D and 2A) are high litter size. Does No. (8D, 9D, 16B and 4A) are lowest litter size. Z for Zaraibi, B for Baladi, A for Alpine and D for Damascus.
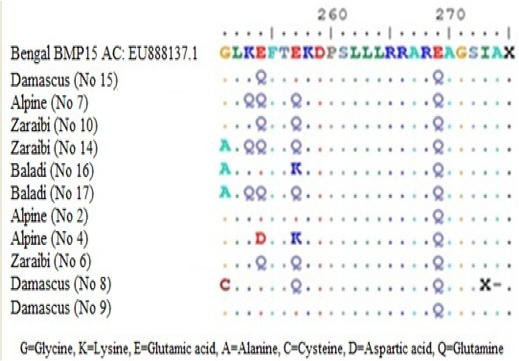
Figure 3: Amino acid alignment showing altered amino acid in four goat breeds under study. Does No (10Z, 6Z, 14Z, 17B, 7A, 15D and 2A) are high litter size. Does No (8D, 9D, 16B and 4A) are low litter size. Z for Zaraibi, B for Baladi, A for Alpine and D for Damascus.
Dna Sequencing, Screening of Snps and Changed Amino Acids
Sequence analysis of BMP15 (141 pb) of 11 females represent the four goat breeds (seven high and four low) and their alignment was shown in Figure 2. The changed amino acids were presented in Figure 3. In the present study, the BMP15 mutation was investigated in the some Egyptian goat breeds. Sequence alignment of all does indicate the existence of seven SNPs among the studied goat breeds having high or low litter size (Table 1).
The results revealed that, there were three SNPs in Zaraibi does number 6, 14 and 10 of high litter size (4, 4 and 3 kids/litter) respectively. There were two SNPs at nucleotide number 760 and 769 G>C and the two corresponding amino acid number 254 and 257 Glu>Gln (Figure 3 and Table 1). These two SNPs can be used as a marker-assisted selection for improvement of litter size in Zaraibi breed.
Moreover, in the Baladi doe number 17 (with 3 kids) has five SNPs at nucleotide number 752 G >C, 757 A>C, 760 G>C, 762 G>A, and 769 G>C showed in Table (1) and the four corresponding amino acids were changed, amino acid number 251 Gly>Ala, 252 Lys>Gln, 254 and 257 Glu> Gln (Figure 3). Also, the Alpine doe number 7 (with 2 kids) had the same previous four SNPs except SNP no. 752. These four SNPs can be used as a marker-assisted selection for high litter size in Baladi and Alpine breeds. In Damascus breed, doe number 15 (with 2 kids) has only one SNP at nucleotide number 760 G>C which leads to change its corresponding amino acids 254 Glu>Gln (Figure 3). From the obtained results there are many SNPs which can use as a marker assisted selection for improving litter size. The SNP in nucleotide 760 is common in all studied breeds (Table 1).
On the other hand, in low litter size does there is many SNPs within each breed. Nucleotides number 752 and 769 in Baladi doe number 16 changed from G>C and G>A and lead to change amino acid 251 Gly>Ala and 257 Glu>Lys. In Alpine breed, SNPs number 762 G>T and 769 G>A were detected in doe number 4 and lead to change translated amino acid 254 Glu>Asp and 257 Glu>Lys (Table I). While, Damascus doe number 8 showed two SNPs, 751 G>T and 769 G>C in Table 1 which lead to change their corresponding amino acids from 251 Gly>Cys and 257 Glu>Gln respectively (Figure 3).
DISCUSSION
PCR-RFLP is a rapid, simple and exact technique for single nucleotide polymorphism (SNP) genotyping. This approach has been used previously to genotype prolific sheep and goat by several research groups (Souza et al., 2001; Davis et al., 2002; Kumar et al., 2006; Guan et al., 2007; Polley et al., 2009).
In our study, PCR-RFLP and SNP approach were used to detect polymorphism in BMP15. PCR-RFLP where used HinfI restriction enzyme to digest the amplified fragment but this enzyme doesn’t digest the DNA amplified fragment and no differences showed between does under study. Similarly, Shokrollahi, (2015) reported that all studied individuals of Markhoz goats showed wild type allele (uncut) with 141 bp in length by using HinfI restriction enzyme.
Seven different SNPs has been detected in does under study. About three SNPs for high litter size does and three for low litter size and one SNP as specific for local Egyptian breeds. So, SNPs which were detected in BMP15 can be used as marker assisted selection (MAS) for high litter size as well as low litter size doe within the breeds. Where, nucleotides number 760 G>C in all high litter size does for all breeds under study (Zaraibi, Baladi, Alpine and Damascus). Furthermore, nucleotide number 757 A>C, 762 G>A, and 769 G>C in high litter size does for three breeds (Alpine, Zaraibi and Baladi). From all results, SNPs considered as a marker assisted selection (MAS) for BMP15 gene which may be improving the litter size in these goat breeds. Moreover, these SNPs affected on amino acids translation where nucleotides number 757, 762 and 769 changed in (Zaraibi, Alpine, Baladi and Damascus) high litter size does lead to change in their corresponding amino acids 253, 254 and 269. Similarly, Nawaz et al. (2013) identified six novel polymorphic sites of BMP15 gene in two goat breeds (Teddy and Beetal) of Pakistan and concluded that BMP15 gene is the major gene which increases the fecundity in Teddy goat. Several studies reported that the variations in BMP15 genes improve the ruminant reproduction and increase the sheep breeding system (Galloway et al., 2000; Hanrahan et al., 2004; McNatty et al., 2005). Another study indicated that BMP15 is the major gene affected prolificacy in Jining Gray goat breed where they detected two point mutations linked with higher prolificacy (Chu et al., 2007).
Not only select high litter size but also we investigated low litter size does within three breeds. Baladi breeds doe number 16 (low litter size) nucleotide number 769 changed from G>A while in high litter size doe (number 17) the same nucleotide (769) changed from G>C with change in amino acid translation 257 Glu>Lys (Doe number 16), so these SNPs can be consider as MAS for low litter size within Baladi breeds.
Within Alpine breed (does number 7 and 4) nucleotide number 762 changed G>A in high litter size does and changed C>T in low litter size doe. Also, Damascus breed (doe) number 8 nucleotide number 751 and 769 changed G>T and G>C in low litter size only while nucleotide number 760 (doe) number 15) changed G>C in high litter size. Also, these SNPs leads to change their corresponding amino acids. So, SNPs detected in BMP15 can be used as marker assisted selection (MAS) for low litter size. The substitutions of the amino acid might lead to a change in protein structure, thus affecting the signaling pathwayduring follicle differentiation and ovulation. Additionally, Cai-lan et al. (2007) reported that exon 2 of BMP15 have a mutation in two nucleotides lead to changes two amino acids in high prolificacy breed (Jining Gray goat) and low prolificacy breed (Inner Mongolia Cashmere).
CONCLUSION
We concluded that, there one main novel SNP present in all high litter size of four breeds and three main novel SNPs detected in does (high litter size) of three breeds (Zaraibi, Alpine and Baladi) leads to change their corresponding amino acids. So, we suggest that the obtained SNPs in BMP15 can be used as MAS for both high and low litter sizes in goats under study.
ACKNOWLEDGEMENTS
We are thankful to all members and assistants of Sakha Animal Production Research Station, Kafr El Sheikh governorate, Egypt for providing necessary facilities to getting samples and data of goat to accomplish this study.
CONFLICT OF INTEREST
There is no conflict of interest.
Authors contribution
All authors contributed equally to carry out this work.
REFERENCES





