Advances in Animal and Veterinary Sciences
Research Article
Isolation of Bacterial Pathogens Associated with Broiler Mortality in Kolar
Kamal Hasan1, Doddamane Rathnamma1, Hogalagere Doddappaiah Narayanaswamy2, Venkataramaiaha Malathi2, Saurabh Gupta3, Shoor Vir Singh3
1Department of Veterinary Microbiology, Veterinary College, Hebbal, Karnataka Veterinary Animal and Fisheries Sciences University (KVAFSU), Bengaluru-560024; 2Veterinary College, Hebbal, Karnataka Veterinary Animal and Fisheries Sciences University (KVAFSU), Bengaluru-24; 3Division of Animal Health, Central Institute for Research on Goats (CIRG), Makhdoom, PO-Farah, Mathura-281122, Uttar Pradesh, India.
Abstract | Tissue samples (livers and ovaries) were collected from thirty birds suffering from dehydration, pyrexia and outbreak of sudden death in a local poultry farm situated at Kolar, India. The outbreak had taken a toll of 180 broiler parent stock birds. Main symptoms and lesions were sudden death and air sacculitis. In order to investigate the causative agents, microbiological examination confirmed 15 samples positive Escherichiaceae coli, 10 (33%) for Salmonella spp. 3 (10%) for Pseudomonas aeruginosa and 2 (6.6%) for Klebsiella spp. On the basis of antibiotic sensitivity tests, Gentamicin showed 100% sensitivity and Doxycycline showed 36.6% sensitivity for the growth inhibition of E.coli isolates. However, Ciprofloxacin and Enrofloxacin shown 100% resistant, which raise serious concerns. This resistance might be due to indiscriminate use of these antibiotics irrespective of etiological agents. Salmonella spp. isolates are highly sensitive to Gentamicin followed by Doxycyclline. Due to involvement of E. coli and Salmonella in public health significance, findings propose control use of antibiotics and only after antibiotic screening.
Keywords | Broilers, Outbreak, Antibiotic sensitivity test, Salmonella spp., Zoonoses
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | May 05, 2017; Accepted | July 01, 2017; Published | July 28, 2017
*Correspondence | Shoor Vir Singh, Division of Animal Health, Central Institute for Research on Goats (CIRG), Makhdoom, PO-Farah, Mathura-281122, Uttar Pradesh, India; Email: shoorvir.singh@gmail.com
Citation | Hasan K, Rathnamma D, Narayanaswamy HD, Malathi V, Gupta S, Singh SV (2017). Isolation of Bacterial Pathogens Associated with Broiler Mortality in Kolar. Adv. Anim. Vet. Sci. 5(7): 312-315.
DOI | http://dx.doi.org/10.17582/journal.aavs/2017/5.7.312.315
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2017 K-Hassan et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Food borne diseases are of serious concern with respect to public health concern, pathogens such as Escherichiaceae coli and Salmonella spp. are among the most frequently isolated bacterial agents of food borne disease outbreaks. Due to its significant morbidity and mortality rates, salmonellosis causes risks to human health (Vo et al., 2014). Currently, poultry farming is rapidly growing agri business in India. Fowl typhoid (FT) caused by Salmonella gallinarum, is an acute septicaemic disease of poultry. The disease is generally associated with high mortality in chicks. Although, there are 2541 known serovars of Salmonella however in India, Salmonella typhimurium and Salmonella enteritidis are the two most common serotypes identified in reported cases of salmonellosis from different sources (Selvaraj et al., 2010). At present, the growth of broiler industry is a serious matter of concern for poultry production. Beef and chicken meat contaminated with fecal organisms may considered essential food hygiene problem particularly enterobacteriaceae including Salmonella spp, E.coli, Proteus as well as Klebsiella spp (Paterson 2006). E. coli are the primary causative agent of cellulitis, septicemia, and air sacculitis in poultry and Salmonella are the causative agent of pullorum disease, fowl typhoid and fowl paratyphoid (Gomis et al., 1997). Therefore, these are the most significant poultry bacterial pathogen. Due to the increase in chicken eggs and meat consumption, the risk of exposure to various animal origin pathogens such as pathogenic E. coli, Salmonella spp., Campylobacter spp. etc has increased. Antibiotics are extensively used as growth promoters in poultry production or to control infectious disease and abuse are considered to be the most vital selecting force to antimicrobial resistance of bacteria (Moreno et al., 2000). Due to enormous use of antibiotics in the field of veterinary medicine, an increased number of resistant bacterial strains were developed in recent years. The intestinal faecal flora from poultry act as a reservoir for transfer of antimicrobial resistance to human pathogens via food chain. The aim of the current work was establish to isolate the micro organisms from an outbreak of broiler farm and the antibiogram pattern against the isolated organisms to overcome the multidrug resistance.
MATERIALS AND METHODS
Collection of Samples
The tissues were collected based on clinical findings and pathogonomic lesions observed during detailed post mortem examination of poultry at Department of Pathology, Veterinary College, Bangalore. A total of 30 samples which includes 12 Liver samples, 14 intestinal samples and 4 ovaries were collected in sterile containers following aseptic precautions and transported in ice pack to laboratory and stored at 4 0C until further use. Tissues were collected form cases exhibiting perihepatitis, enteritis, air sacculitis, yolk sac infection pneumonitis and pericarditis.
Isolation and Identification
The tissue samples were collected aseptically and used for microbiological test. The samples were inoculated into Brain Heart infusion broth for primary enrichment, then incubate the broth 24 hours at 370C and from broth streaked on MacConkey Agar and Eosin Methylene Blue (EMB) agar plate. The plate was incubated at 370C examined after 24 hours for growth and change in the color of the medium. After overnight incubation the bacterial growth was observed as large pink colonies at MacConkey and Greenish metallic sheen colonies at EMB agar. Both lactose fermenting and non lactose fermenting colonies were found. Salmonella spp. organisms will grow on differential plating media such as MacConkey and SS Agar. Confirmation of Salmonella spp. was done by culturing on selective media such as Xylose lysine deoxycholate (XLD) Agar and observation of colony characteristics such as black centered pale pink colony respectively. Various biochemical tests were performed for species identification. For this study isolated organisms with supporting growth characteristic of E. coli on EMB and Salmonella on XLD were subjected to various biochemical tests named TSI agar slant reaction, MR–VP, Indole reaction and citrate utilization test were carried out for identification of suspected organisms. All the isolates from different sources were tested for the detection of Escherichiaceae coli, Salmonella spp., Pseudomonas spp. and Klebsiella spp. (Cruickshank, 1980).
Antimicrobial Sensitivity Pattern of the Isolated Salmonella and E. coli
On the basis of colonial morphology, positive isolates were finalized for biochemical tests. Then isolated organisms were screened for anti-microbial sensitivity by using standard Kirby Bauer disc diffusion method. The following antibiotics and disc potencies were used: GEN: Gentamicin (10μg), DO: Doxycycline (30μg), CIP: Ciprofloxacin (5μg), ENR: Enrofloxacin (5μg), AMX: Amoxicillin (10μg), N: Norfloxacin (10μg), CL: Colistin (10μg), K: Kanamycin (30µg) from HIMEDIA Ltd (Mumbai, India).The overnight nutrient broth cultured Salmonella and E.coli isolates were poured on Meuller Hinton agar and spread uniformly with the help of sterile glass spreader. Antibacterial discs were applied aseptically to the surface of the plate at an appropriate distance with the help of sterile forceps and incubated at 37˚C for 24 hours, aerobically. Antibiotic sensitivity pattern of isolated E. coli and Salmonella were performed against 14 commonly used antibiotics belonging to different groups . Using a metric ruler, measured the diameter of the zone of inhibition (if present) for each antibiotic used. By comparing the measurement obtained from the individual antibiotics to the table of standards to determine if the bacterial species tested is resistant or sensitive to the antibiotic (Bauer et al., 1966).
RESULTS
In the present study, out of the total 30 samples collected, E. coli were recovered from fifteen (50%) samples, three were Pseudomonas spp. Ten individual colonies of Salmonella spp., 2 colonies were from Klebsiella spp. were isolated (Figure 1). The gram negative isolates were confirmed by various biochemical tests (Table 1). E.coli isolates, were 100 % sensitive to gentamicin and 100 % resistant to ciprofloxacin, enrofloxacin and In case of Colistin, 12 isolates were resistant and 3 isolates were sensitive and In case of doxycycline, 9 isolates were resistant, 6 isolates were sensitive. Antimicrobial resistant pattern in norfloxacin showed 3 isolates were resistant and 12 were sensitive. In case of kanamycin, 5 isolates were resistant,4 were sensitive and 6 were intermediate sensitive. In case of amoxicillin 13 isolates were resistant, 1 was sensitive and 1 was intermediate sensitive (Table 2). Out of 30 isolates obtained , Majority were E.coli isolates (50%) followed by Salmonella spp.(33.3%), Pseudomonas spp.(10%), and Klebsiella spp.(6.6%) (Table 4).
The Salmonella spp. isolates, 100 % sensitive to gentamicin and 100 % resistant to ciprofloxacin, enrofloxacin and amoxycillin. In case of colistin, 7 isolates were resistant and 3 isolates were sensitive and In case of doxycycline, 2 iso-
Table 1 : Biochemical tests’ results of gram-negative isolate
|
Isolates |
Motility |
C |
O |
NR |
I |
MR |
VP |
Citrate |
Urease |
TSI |
||
|
Slant/Butt |
Gas |
H2S |
||||||||||
|
Escherichia coli (n=15) |
Motile |
+ |
- |
+ |
+ |
- |
- |
- |
- |
Y/Y |
+ |
- |
|
Pseudomonas species (n=3) |
Motile |
+ |
+ |
+ |
- |
- |
- |
+ |
+ |
P/P |
- |
- |
|
Klebsiellaspecies (n=2) |
Non-motile |
+ |
- |
+ |
- |
- |
+ |
+ |
+ |
Y/Y |
+ |
- |
|
Salmonella species (n=10) |
Motile |
+ |
- |
+ |
- |
- |
+ |
+ |
+ |
Y/Y |
+ |
- |
*C- Catalase test; O- Oxidase test; NR- Nitrate reduction test; I- Indole production; MR- Methyl-red test; VP- Voges-Proskauer test; Citrate- Citrate utilization test; Urease- Urease activity; TSI- Triple sugar iron fermentation; Y- yellow/acidic; P- pink/alkaline; V- Variable; ‘+’: Positive; ‘-’: Negative.
Table 2 : Antimicrobial resistance pattern of E.coli isolates
|
Sno |
GEN |
CL |
CIP |
DO |
N |
K |
ENR |
AMX |
|
1 |
S |
R |
R |
S |
S |
R |
R |
R |
|
2 |
S |
S |
R |
S |
S |
R |
R |
R |
|
3 |
S |
R |
R |
S |
S |
R |
R |
R |
|
4 |
S |
R |
R |
R |
R |
R |
R |
I |
|
5 |
S |
S |
R |
S |
R |
R |
R |
R |
|
6 |
S |
S |
R |
S |
S |
S |
R |
R |
|
7 |
S |
R |
R |
S |
R |
S |
R |
R |
|
8 |
S |
R |
R |
R |
S |
I |
R |
R |
|
9 |
S |
R |
R |
R |
S |
I |
R |
R |
|
10 |
S |
R |
R |
R |
S |
R |
R |
R |
|
11 |
S |
S |
R |
R |
S |
R |
R |
R |
|
12 |
S |
S |
R |
R |
S |
I |
R |
S |
|
13 |
S |
S |
R |
R |
S |
I |
R |
R |
|
14 |
S |
S |
R |
R |
S |
I |
R |
R |
|
15 |
S |
S |
R |
R |
S |
I |
R |
R |
Table 3: Antimicrobal resistance against Salmonella isolates
|
Sno |
GEN |
CL |
CIP |
DO |
N |
K |
ENR |
AMX |
|
1 |
S |
R |
R |
S |
S |
R |
R |
R |
|
2 |
S |
S |
R |
S |
S |
R |
R |
R |
|
3 |
S |
R |
R |
S |
S |
R |
R |
R |
|
4 |
S |
R |
R |
R |
R |
R |
R |
R |
|
5 |
S |
S |
R |
S |
R |
R |
R |
R |
|
6 |
S |
S |
R |
S |
S |
S |
R |
R |
|
7 |
S |
R |
R |
S |
R |
S |
R |
R |
|
8 |
S |
R |
R |
S |
S |
I |
R |
R |
|
9 |
S |
R |
R |
S |
S |
I |
R |
R |
|
10 |
S |
R |
R |
R |
S |
R |
R |
R |
lates were resistant, 8 isolates were sensitive. Antimicrobial resistant pattern in norfloxacin showed 3 isolates were resistant and 7 isolates were sensitive. In case of kanamycin, 6 isolates were resistant, 2 isolates were sensitive and 2 isolates were intermediate sensitive (Table 3).
DISCUSSION
The important Zoonoses bacteria isolated from broiler meat in the present study were Escherichiaceae coli, Salmonella spp. Pseudomonas aeruginosa and Klebsiella spp. Thirty isolates were obtained which were confirmed by biochemical tests. This in agreement with findings of El Sayed et al. (2017) stated that the E.coli and Salmonella spp. were the predominant isolated bacteria from outbreaks of poultry diseases.
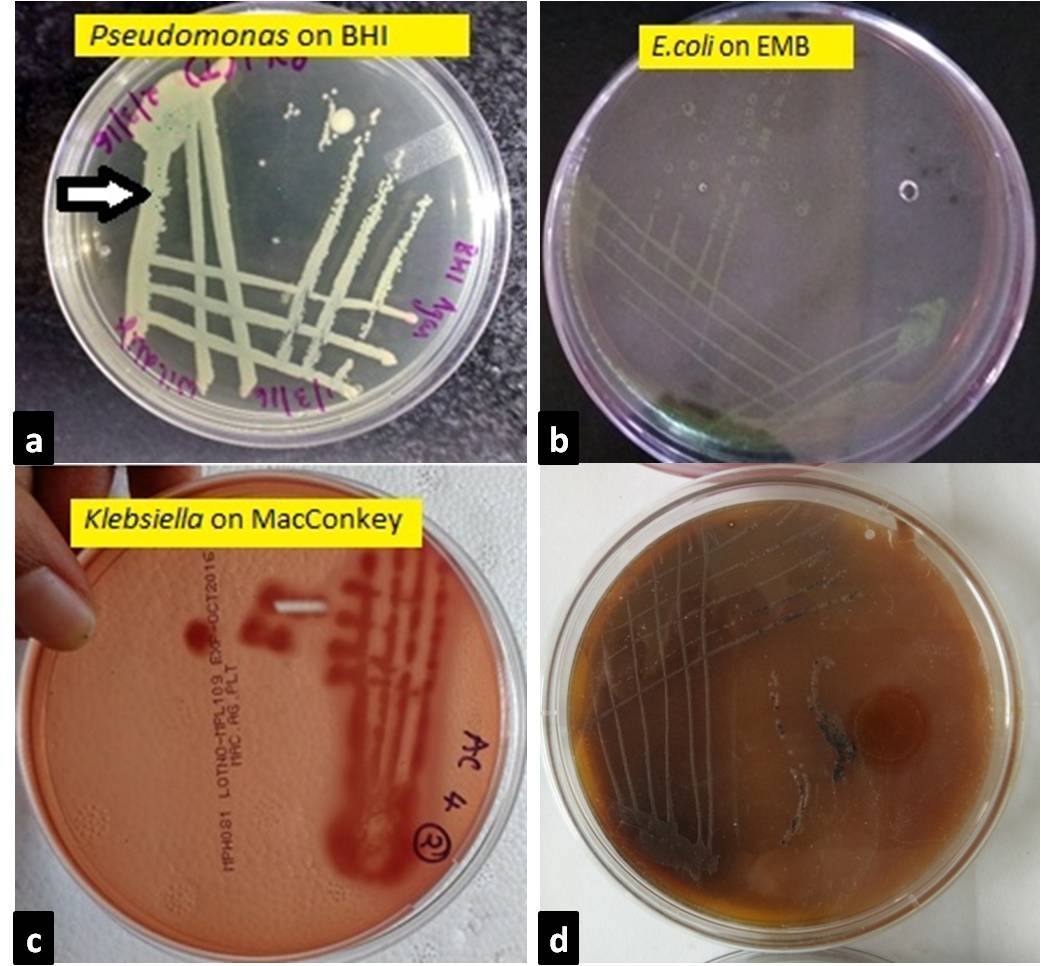
a. Pseudomonas colonies appeared greenish blue on BHI agar after 24 hr incubation; b. On EMB agar after 24hr incubation, E.coli colonies appeared as bluish black with a characteristic appearance of metallic sheen; c. On MacConkey agar after 24hr incubation, Klebsiella colonies appeared as large, pink and mucoid indicating lactose fermentation; d. On XLD agar, Blackish colonies indicating salmonella
Table 4 : Percentage of different bacterial isolates obtained
|
Isolates |
Positive |
Percentage of Positive Isolates |
|
Escherichia coli |
15 |
50 % |
|
Salmonella |
10 |
33 % |
|
Pseudomonas |
03 |
10 % |
|
Klebsiella |
02 |
6.6 % |
E.coli isolates were 100 % sensitive to gentamicin which is in agreement with the Ameen et al. (2017) stated that highest level of sensitivity was found against gentamycin antibiotic .The resistance of all the E. coli isolates and Salmonellaspp. isolates procured during the study period against commonly used antibiotics such as ciprofloxacin and enrofloxacin is a matter of serious concern. This resistance might be due to indiscriminate use of these antibiotics irrespective of aetiological agents (Kumar et al., 2013) as well different drug interaction due to concurrent administration is performed irrespective of type of drug and their interaction (Rahal et al., 2007).
Conclusions
Isolation of E. coli, Salmonellaspp., Pseudomonas aeruginosa and Klebsiella spp is one of the most important food borne pathogens of public health importance. Hence, special emphasis need to be given for judicious selection of antibiotics, preferably after antibiotic sensitivity testing and judicious use of such antibiotics at an optimum dose for sufficient duration to ensure effective treatment and control of various diseases caused by different organisms in poultry.
ACKNOWLEDGEMENTS
Authors are thankful to NAE project ‘Animal Disease Registry and Tissue Bank’, Department of Veterinary Pathology, Veterinary College, Hebbal, Bengaluru.
CONFLICT OF INTEREST
We declare that we have no conflict of interest.
AUTHORS CONTRIBUTION
All authors contributed equally
REFERENCES