Advances in Animal and Veterinary Sciences
Research Article
Molecular and Phenotypic Characterization of Antimicrobial Resistance in Gram Negative Bacteria Recovered from Subclinical Mastitis
Gamal Younis, Amal Awad*, Nour Ashraf
Department of Bacteriology, Mycology and Immunology; Faculty of Veterinary Medicine; Mansoura University; Mansoura, 35516, Egypt.
Abstract | This study was undertaken to estimate the prevalence and antimicrobial susceptibility of gram negative bacteria (GNB) recovered from dairy cows with subclinical mastitis (SCM) with detection of β-lactamase encoding genes, sulphonamide encoding gene and intergron classes by polymerase chain reaction (PCR). In total, 180 cow’s milk samples were collected from three different dairy farms. Milk samples were subjected to physical examination and subsequently to California mastitis test (CMT). Subclinically infected milk samples were subjected to bacterial isolation and GNB isolates were subsequently tested for their antimicrobial susceptibility to 11 antimicrobial agents by disc diffusion method. All GNB isolates were investigated for the presence of integron classes, β-lactamase encoding genes (blaTEM, blaSHV, blaCTX) and sulphonamide encoding gene (sul1) gene by using PCR.The overall prevalence of subclinical mastitis (SCM) in lactating dairy cows was 55.56% (100/180). Bacteriological analysis revealed the presence of gram negative bacteria in 35% (35/100) of the tested samples. E. coli was found to be the most prevalent organism (10) followed by Klebsiella pneumonia (5), Proteus mirabilis (4) Enterobacter aerogenes (3), Serratia liquefaciens (3), Providencia rettgeri (2), Proteus vulgaris (2),Citrobacter freundii (2), Citrobacter diversus (1), Enterobacter agglomerans (1), Enterobacter cloacae (1) and Yersinia enterocolitican (1). Antimicrobial susceptibility results revealed that the highest resistance was observed for amoxicillin, clindamycin, trimethoprim/sulfamethoxazole, vancomycin and rifapime. All bacterial isolates revealed a multidrug resistance (MDR). By PCR 100%, 80% and 60% of gram negative isolates harbored blaTEM, blaSHV and blaCTX , respectively. Meanwhile, sul1 was detected in 80% (28/35) of the tested isolates. Class 1 intergron was detected in 91.4% (32/35) and Class 2 intgrons could not be identified in all the tested isolates. This study indicates the need for effective control measures to challenge the increase in occurrence of subclinical mastitis. The overall proportion of antimicrobial resistance was high. As a result, this study suggests that the risk of dissemination of resistance gram negative bacteria through the food chain.
Keywords | Dairy farm, β-lactamase, Gram negative, Integrons, SCM, Sul1 gene.
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | February 15, 2017; Accepted | April 14, 2017; Published | April 21, 2017
*Correspondence | Amal Awad, Department of Bacteriology, Mycology and Immunology; Faculty of Veterinary Medicine; Mansoura University; Mansoura, 35516, Egypt; Email: amalabdo@mans.edu.eg
Citation | Younis G, Awad A, Ashraf N (2017). Molecular and phenotypic characterization of antimicrobial resistance in gram negative bacteria recovered from subclinical mastitis. Adv. Anim. Vet. Sci. 5(5): 196-204.
DOI | http://dx.doi.org/10.17582/journal.aavs/2017/5.5.196.204
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2017 Younis et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Mastitis is consider the common infectious disease affects dairy cattle and cause numerous losses on the dairy production. Large part of this cost is not related to cows with clinical mastitis but it is the result of sub clinically mastitic cow with consequent decrease in milk production (Ott and Novak, 2001). Clinical mastitis has apparent symptoms but SCM has concealed symptoms most farmers are ignorant of it. SCM has direct and indirect significances which causes serious economical loss for the farmers (Godkin et al., 1990). The major economic losses are caused by decrease in milk production, premature culling and treatment costs which account for 78, 14 and 8% of the total loss, respectively (Scheprs and Dijkhuizen, 1991). Another significant economic loss is due to extra work for the farmer.
Coliforms, i.e. E. coli and Klebsiella spp., are natural intestinal flora of the bovine (Sandholm and Pyörälä, 1995). It is spread and contaminates the environment through faeces, and contaminates bovine udder and subsequently results in clinical and subclinical mastitis.
Multiple drug resistance of mastitis pathogens has been stated worldwide (Waller et al., 2011; Chaudhary and Payasi, 2013). This is because of impromptu use of the antibiotics by farmers either to improve animal performance or in treatment. The emergence of MDR especially those exhibiting resistance to cephalosporins due to production beta- lactamases has attracted attention worldwide. Antimicrobial resistance mechanism can be spread by genetic mutation which occur at low rate and acquistation to a lot of genes that mediate the resistance to their host microorganism, this acquistation of resistant gene is the main contributor for spreading of antimicrobial resistance and this occur either by horizontal transfer or vertical transfer, the horizontal transfer include: mobile genetic parts such as plasmid and transpons (Xu et al. 2012), integron also have been documented to play major role in spreading of the resistant genes (Hall and Collis, 1995; Mazel, 2006).
The purpose of this study was to investigate the prevalence of SCM in dairy farms in Dakahlia governorate, Egypt. Furthermore, to identify gram negative pathogens, determine their antibiotic susceptibility phenotypes with detection of β-lactamase encoding genes, sulphonamide encoding gene and integron classes.
Materials and methods
Samples Collection
A total of 180 milk samples were collected from 3 different dairy farms located in the district of Mansoura city, Dakahlia,Egypt. 15 randomly selected animal from each farm, four samples were collected from the 4 quarters of each animal with no signs of clinical mastitis. Milk samples were tested physically and with CMT to reveal subclinical mastitis prevalence. The samples were collected under aseptic condition,briefly; the udder was washed with warm water, dried and the teat disinfected with 70% alcohol. About15 ml discarded from the fore milk and the next 15 ml collected into screw cap bottles and transported to the laboratory on ice box. Samples were stored at 4ºC from the time of collection until time of examination.
Bacteriological Examination
Bacterial culture and isolation were performed at the Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Mansoura University, Egypt. For bacteriological examinations, Milk samples were centrifuged for 15 minutes at 3000 rpm and a loopful was sampled from the sediment and inoculated on MacConkey’s agar. The inoculated plates were then incubated at 37◦C for 24 and 48hr and the isolates were identified conventionally according to (Edwards and Ewing, 1986).All gram negative bacterial isolates were confirmed biochemically using API 20E system (BioM´erieux, Marcy-l’´Etoile, France)
Antimicrobial Susceptibility Testing:
All bacterial isolates were screened for their antibiotic susceptibility against 11 antibiotic agents by disc diffusion method on Mueller-Hinton agar (Oxoid Ltd.) using the following disks (Oxoid) amoxacillin (AX; 25μg),doxytetracyclin (DO; 30μg), Rifapime (RA; 5 μg), chloramphenicol (C; 30 μg), azithromycin (AZM; 15μg), sulphamethoxazole-trimethoprim (SXT; 25μg), gentamycin (CN;10μg), levofloxacin(LEV; 10 μg), norfloxacin (NOR; 10μg),vancomycin (VA; 30μg), clindamycin( DA; 10 μg). The inhibition zones were scored as sensitive, intermediate susceptibility and resistant according to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2010). The isolates were defined as multidrug-resistant if they exhibited resistance to three or more different antimicrobials classes (Magiorakos et al., 2012).
Molecular Characterization of Antibiotic Resistant Genes
Bacterial DNA for PCR analysis was prepared using a Genomic DNA purification kit QIAamp DNA Mini Kit Catalogue no.51304 according to the manufacturer’s instructions. The presence of genes associated with resistance to sulfonamides (sul1), beta-lactams (blaSHV, blaTEM, blaCTX) were determined by PCR and the set of primers from Metabion (Germany), used for each gene is shown in Table 1. Preparation of PCR Master Mix according to Emerald Amp GT PCR mastermix (Takara) Code No. RR310A kit: PCR reactions were performed in a total volume of 25 µl, including template DNA (6μl), forward and reverse primers (1μl), 12.5μl of Emerald Amp GT PCR mastermix (2x premix) and 4.5μl of PCR grade water. Amplification reactions were carried out using Applied Bio systems 96-well thermal cycler. Temperature and time conditions of each primer during PCR are demonstrated in Table 2. Electrophoresis of amplified products was carried out using 1.5% agarose gel stained with ethidium bromide and detected by UV transillunination. Amplified genes were identified on the basis of fragment size.
Screening for Class 1 and Class 2 Integrons
Genomic DNA was extracted using QI Aprep Spin Miniprep Kits Catalogue no. 27104 following the user instruction. PCR amplification of classes 1 and 2 integrase genes was performed in 25 μL reaction mixtures including, 1 μl of each primer (Metabion, Germany) (Table 1), Template DNA (6μl), 12.5μl of Emerald Amp GT PCR mastermix (2x premix) and 4.5μl of PCR grade water,. Amplifications were performed in a Thermal Cycler (Applied Bio systems 96-well) using the program mentioned in Table 2. The amplified PCR products were separated by electrophoresis in 1% agarose gels and visualized after staining with ethidium bromide.
Table 1: Primers sequence used in PCR assay
|
Primer |
Sequence |
Amplified product |
Reference |
|
blaTEM |
ATCAGCAATAAACCAGC |
516 bp |
Colom et al., (2003) |
|
CCCCGAAGAACGTTTTC |
|||
|
blaSHV |
AGGATTGACTGCCTTTTTG |
392 bp |
|
|
ATTTGCTGATTTCGCTCG |
|||
|
Sul1 |
CGGCGTGGGCTACCTGAACG |
443 bp |
Sabarinathet al., 2011 |
|
GCCGATCGCGTGAAGTTCCG |
|||
|
Int1 |
CCTCCCGCACGATGATC |
280 bp |
Kashif et al., (2013) |
|
TCCACGCATCGTCAGGC |
|||
|
Int2 |
TTATTGCTGGGATTAGGC |
250 bp |
|
|
ACGGCTACCCTCTGTTATC |
|||
|
blaCTX |
ATG TGC AGY ACC AGT AAR GTK ATG GC |
593 bp |
Archambault et al., (2006) |
|
TGG GTR AAR TAR GTS ACC AGA AYC AGC GG |
Results
A total of 180 milk samples from 3 different dairy farms were tested. Of these, 55.56% (100/180) affected with subclinical mastitis after testing by CMT. A total of 35 gram negative bacterial isolates have been recovered including, E. coli 28.57% (10/35) followed by K.pneumonia 14.28% (5/35), P. mirabilis 11.42% (4/35) S. liquefaciens 8.57% (3/35), E. agglomerans 2.85%(1/35), E. aerogenes 8.57% (3/35), C. diversus 2.85% (1/35), C. freundii 5.7% (2/35), E. cloacae 2.85% (1/35), ,P. retttgeri 5.7% (2/35),, P. vulgaris 5.7% (2/35), Y. enterocolitica 2.85% (1/35) (Table 3).
Antimicrobial susceptibility tests were conducted on the 35 gram negative bacterial isolates identified in this study. Variable resistance patterns were seen among the used antibiotics (Table 4). Bacterial isolates were showing a high resistance to amoxicillin 35(100%), clindamycin 31(88.75%), rifapime 28(80%), trimethoprim/sulfamethoxazole 26 (74.3%), vancomycin 24(68.6%), a moderate resistance to azithromycin18(51.4%), gentamycin 17(48.6%), doxycycline 17(48.6%), chloramphenicol 13(37.1%) and a low resistance to norofloxacin 6(17.14%) and levofloxacin 3(8.57%) (Table 3). All bacterial isolates revealed MDR.
Genetically, all 35 gram negative bacterial isolates were screened for the presence of β-lactamase encoding genes (blaTEM, blaSHV, blaCTX) and sulfonamide encoding gene (sul1). The detection rate of β-lactamases resistance genes
Table 2: Cycling conditions of the different primers during PCR
|
Gene |
Primary denaturation |
Secondary denaturation |
Annealing |
Extension |
No. of cycles |
Final extension |
|
blaTEM |
94˚C 5 min. |
94˚C 30 sec |
54˚C 45 sec |
72˚C 45 sec |
35 |
72˚C 10 min. |
|
blaSHV |
94˚C 5 min. |
94˚C 30 sec |
54˚C 45 sec |
72˚C 45 sec |
35 |
72˚C 10 min. |
|
sul1 |
94˚C 5 min. |
94˚C 30 sec. |
60˚C 45 sec |
72˚C 45 sec |
35 |
72˚C 10 min. |
|
Int2 |
94˚C 5 min. |
94˚C 30 sec. |
50˚C 30 sec. |
72˚C 30 sec. |
35 |
72˚C 7 min. |
|
Int2 |
94˚C 5 min. |
94˚C 30 sec. |
48˚C 30 sec. |
72˚C 30 sec. |
35 |
72˚C 7 min. |
|
blaCTX |
94˚C 5 min. |
94˚C 30 sec |
60˚C 45 sec |
72˚C 45 sec |
35 |
72˚C 10 min. |
Table 3: Distribution of different bacteria isolates in cow’s milk samples.
|
Bacteria |
Number |
Percentage |
|
E. coli |
10 |
28.57% |
|
Klebsiella pneumonia |
5 |
14.28% |
|
Proteus mirabilis |
4 |
11.42% |
|
Providencia rettgeri |
2 |
5.7% |
|
Proteus vulgaris |
2 |
5.7% |
|
Serratia liquefaciens |
3 |
8.57% |
|
Enterobacter agglomerans |
1 |
2.85% |
|
Yersinia enterocolitica |
1 |
2.85% |
|
Citrobacter freundii |
2 |
5.7% |
|
Enterobacter cloacae |
1 |
2.85% |
|
Enterobacter aerogenes |
3 |
8.57% |
|
Citrobacter diversus |
1 |
2.85% |
|
Total |
35 |
100% |
was higher; blaTEMwas detected in all gram negative strains, blaSHV was detected in 80% (28/35) meanwhile blaCTX was found in 60% (21/35).sul1 was detected 28 (80%) isolates. Regarding to integron classes, Class 1 integron was detected in 91.4% (32/35) and Class 2 integrons could not be identified in all the gram negative isolates.
Discussion
In this study the prevalence of subclinical mastitis is 55.56% (100/158) this high prevalence agrees with an earlier report by (Chye et al., 2004; Marimuthu, 2014) who suggested that the high prevalence of subclinical mastitis was possibly as a result of unhygienic milking practices, In our study we focused on gram negative bacteria recovered
from examined samples. a variety of gram negative bacterial spp. have been recovered from cow’s milk samples,E.coli was the most common in between gram negative bacteria isolates 10 (28.57%). These results agreed with several previous studies (Gianneechini, 2002; Makovee and Ruegg, 2003; Malinowski, 2006). The high incidence of E.coli that cause mastitis in our study may be due to post milking teat dipping and the usual use of drugs in dry period, which effect on contagious Gram positive bacteria not on environmental one such as E.coli (Fang and Pyoralai, 1996).
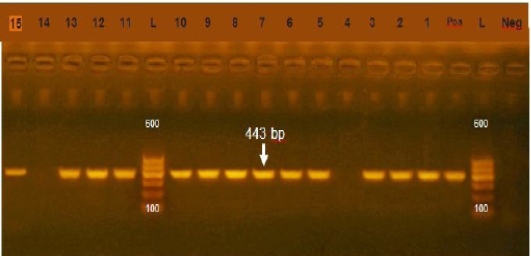
Figure 1: Agarose gel electrophoresis showing amplification of 443bp fragment using sul1 primer. (Lane 1,2,3,5-13,15) positive, Lane 4,14: negative, Neg: negative control, Pos: positive control, L: Ladder 600bp
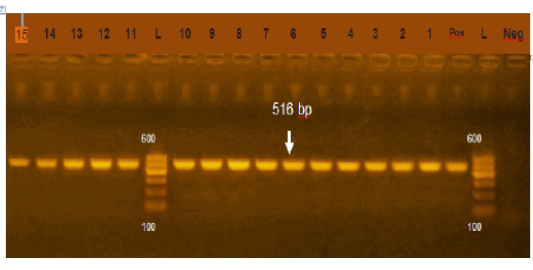
Figure 2: Agarose gel electrophoresis showing amplification of 516bp fragment using bla TEM primer. Lane 1-15: Positive, Neg: negative control, Pos: positive control, L: Ladder 600bp
Table 4: Percentage of antimicrobial resistance in bacterial isolates:
|
Antimicrobial drug |
Code |
Antimicrobial class |
Sensitive |
Resist |
|
Gentamycin |
CN |
Aminoglycosides |
18(51.4%) |
17(48.6%) |
|
Levofloxacin |
LEV |
Quinolones |
32(91.4%) |
3(8.57%) |
|
Norofloxacin |
NOR |
Quinolones |
29(82.85%) |
6(17.14%) |
|
Vancomycin |
VA |
Glycopeptide |
8(22.9%) |
27(77.14%) |
|
SXT |
Sulphonamides |
9(25.7%) |
26(74.3%) |
|
|
Doxycycline |
DO |
Tetracyclines |
18(51.4%) |
17(48.6%) |
|
Clindamycin |
DA |
Lincosamides |
4(11.4%) |
31(88.75%) |
|
Amoxicillin |
AX |
ß-Lactam |
0.00 |
35(100%) |
|
Rifapime |
RA |
Rifamycin |
7(20%) |
28(80%) |
|
Chloramphenicol |
C |
Phenicols |
22(62.85%) |
13(37.1%) |
|
Azithromycin |
AZM |
Macrolides |
17(48.6%) |
18(51.4%) |
Table 5: Antimicrobial resistance profiles and resistance gene pattern of gram negative bacterial isolates.
|
Samples |
Strain |
Integron class 1 |
Integron class 2 |
Antibiotic resistance pattern |
Resistance genes |
|
1 |
E.coli |
+ |
- |
AX, RA, STX, CN, VA. |
TEM, CTX, SUL1 |
|
2 |
E.coli |
+ |
- |
AX, DO,RA,AZM,SXT,CN,NOR,VA, OX |
TEM,CTX,SUL1 |
|
3 |
E.coli |
+ |
- |
AX,RA,AZM,STX,CN,VA. |
TEM,SHV,CTX,SUL1 |
|
4 |
E.coli |
+ |
- |
AX,DO,RA,SXT,CN,LEV,NOR,VA. |
TEM,SHV,CTX,SUL1 |
|
5 |
E.coli |
+ |
- |
AX,DO,RA,C,AZM,SXT,VA. |
TEM,SHV,CTX,SUL1 |
|
6 |
E.coli |
+ |
- |
AX,DO,RA,SXT,CN,RF,NOR,VA,OX |
TEM,SHV,SUL1 |
|
7 |
E.coli |
+ |
- |
AX,C,AZM,SXT,CN,NOR,OX. |
TEM,SHV,SUL1 |
|
8 |
E.coli |
+ |
- |
AX,DO,CN,LEV,VA. |
TEM,SHV,SUL1 |
|
9 |
E.coli |
+ |
- |
AX,SXT,CN,VA. |
TEM,SHV,CTX,SUL1 |
|
10 |
E.coli |
+ |
- |
AX,RA,C,AZM,SXT,VA,OX. |
TEM,SHV,CTX,SUL1 |
|
11 |
K.pneumoniae |
+ |
- |
AX,RA,SXT,CN |
TEM,SHV ,SUL1 |
|
12 |
K.pneumoniae |
+ |
- |
AX,RA,SXT,RF,VA. |
TEM,SHV,CTX,SUL1 |
|
13 |
K.pneumoniae |
+ |
- |
AX,RA,SXT,CN |
TEM,SHV,SUL1 |
|
14 |
K.pneumoniae |
+ |
- |
AX,RA,C,STX,CN,NOR. |
TEM,SHV,CTX,SUL1 |
|
15 |
K.pneumoniae |
+ |
- |
AX,RA,VA. |
TEM,SHV,SUL1 |
|
16 |
P.mirabilis |
+ |
- |
AX,RA,SXT,CN,VA. |
TEM,SHV,CTX,SUL1 |
|
17 |
P.mirabilis |
+ |
- |
AX,RA,RF,OX. |
TEM,SHV,CTX |
|
18 |
P.mirabilis |
+ |
- |
AX,RA,C,CN. |
TEM,SHV,CTX,SUL1 |
|
19 |
P.mirabilis |
+ |
- |
AX,DO,RA,C,AZM,CN,VA. |
TEM,CTX |
|
20 |
S.liquefaciens |
+ |
- |
AX,DO,SXT,RA,C,AZM,VA. |
TEM,SHV,CTX,SUL1 |
|
21 |
S.liquefaciens |
+ |
- |
AX,DO,RA,C, SXT. |
TEM,SHV,CTX,SUL1 |
|
22 |
S.liquefaciens |
- |
- |
AX,RA,SXT,VA. |
TEM,SHV,SUL1 |
|
23 |
E.aerogenes |
- |
- |
AX,DO,C,AZM,SXT,VA,OX. |
TEM,SHV,SUL1 |
|
24 |
E.aerogenes |
- |
- |
AX,RA,AZM,VA,OX. |
TEM,SHV,CTX,SUL1 |
|
25 |
E.aerogenes |
+ |
- |
AX,DO,RA,C,RF,AZM,VA. |
TEM,SHV,SUL1 |
|
26 |
P.rettgeri |
+ |
- |
AX,DO,RA,AZM,SXT,VA |
TEM,CTX,SUL1 |
|
27 |
P.rettgeri |
+ |
- |
AX,AZM,SXT,CN,NOR,OX. |
TEM,SHV,CTX. |
|
28 |
C.freundii |
+ |
- |
AX,DO,RA,C,AZM,SXT,VA, |
TEM, CTX,SUL1 |
|
29 |
C.freundii |
+ |
- |
AX,DO,RA,AZM,SXT,VA. |
TEM,SHV,CTX |
|
30 |
P.vulgaris |
+ |
- |
AX,DO,C,RF,VA. |
TEM,SHV |
|
31 |
P.vulgaris |
+ |
- |
AX,RA,AZM,SXT,VA. |
TEM,CTX,SUL1 |
|
32 |
E. agglomerans |
+ |
- |
AX,DO,RA,AZM,SXT,CN,LEV,VA,OX. |
TEM,SHV,CTX,SUL1 |
|
33 |
E.cloacae |
+ |
- |
AX,DO,RA,AZM,SXT, VA |
TEM,SUL1 |
|
34 |
Y.enterocolitica |
+ |
- |
AX,DO, RA,C,SXT,CN,VA. |
TEM,SHV,SUL1 |
|
35 |
C.diversus |
+ |
- |
AX, AZM,VA. |
TEM,SHV,SUL1 |
In addition, it may be attributed to lower hygienic measures and the high contamination of bedding that result in udder infection come from fecal contamination from the contaminant beddings (Liebiseh et al., 1994) Furthermore K. Pneumoniae was recorded in our study with incidence of 14.28% which is higher than the result reported by (Lalrinthuanga et al., 2003) and lower than (Katsande et al., 2013).
Due to excessive use of antibiotic leads to appearance of multidrug resistance (MRD) phenomenon. Which lead to blocking the ability of antimicrobial agents in treating the infectious diseases (Martinez and Baquero, 2002). The antimicrobial resistance of bacteria is consider one of the most dangerous public health that the animal which carry antibiotic resistance bacteria affect human health as enter human food chain through consuming meat and any other animal products, By water run off from farms and any other pathways (Collignon et al., 2005).
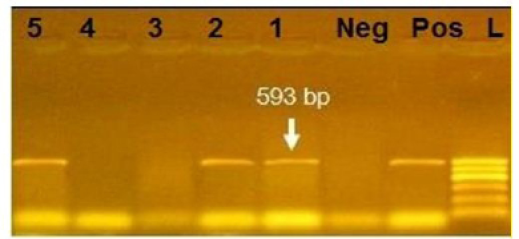
Figure 3: Agarose gel electrophoresis showing amplification of 593bp fragment using bla CTX primer. Lane 1,2,5: Positive. Lane 3,4: negative, Neg: negative control, Pos: positive control, L: Ladder 600bp
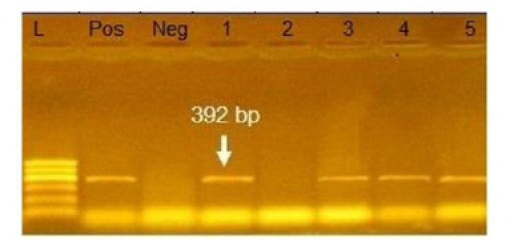
Figure 4: Agarose gel electrophoresis showing amplification of 392bp fragment using bla SHV primer. Lane 1,3,4,5: Positive. Lane 2: negative, Neg: negative control, Pos: positive control, L: Ladder 600bp
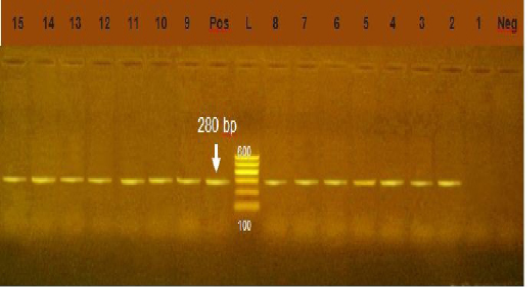
Figure 5: Agarose gel electrophoresis showing amplification of 280bp fragment using int1 primer. Lane 2-15: positive. Lane 1: negative, Neg: negative control, Pos: positive control, L: Ladder 600bp
The multidrug resistant pathogens which have ability to pile up resistance genes are the main cause of defeat in treatment of infectious diseases this lead to increasing rate of morbidity and mortality, and a larger economic loss, for health care apparatus and individuals (Lipsitch et al., 2002). All GNB (35%) isolated were multidrug resistance. Findings of our study are higher than the result of (Hammad and Shimamoto, 2011) who recorded that 4 out a total of 75 isolates (5.3%) only seen to be multidrug resistant.
In many countries the bacteria isolated from bovine mastitis show resistant phenotypes; Even though, there is difference in the resistance patterns of bacterial groups among the countries, which may demonstrate differences in anti-microbial treatment (Bengtsson et al., 2009; Hawari and Al-Dabbas 2008; Lipsitch et al., 2002). The level of resistance of certain antibiotics in GNB this may explained by excessive use of these antibiotics in treatment of animal in Egypt.
Resistance of sulfonamide in gram negative bacteria isolated from human, animals, world wide show high prevalence (Moreno et al., 2006). In our study the prevalence of sul1 encoding gene was 85.7% (Figure 1) which was higher that of (Arabi et al., 2015) who recorded sul1 in 81% of resistant isolates also (Hindi et al., 2013) who detected sul1 in 15% of the examined samples and (Antunes et al., 2005) reported sul1 in 76% of resistant sulfonamides isolates. The resistance to sulfonamides in gram negative bacteria result from acquisition of either two genes of sul1 or sul2 that code (DHPS) which resist this drug. Most multi-resistant gram-negative bacteria harbor Class 1 integrons which carry the sul1 gene (Liu et al., 2009)
Phenotypically, GNB have high resistance to large number of antimicrobials agent used in this study. Particularly, β-lactams, GNB have 100% resistant to amoxicillin and this resistance properties associated with 100%, 80% and 60% prevalence of blaTEM, blaSHV and blaCTX (Figure 2, 3 and 4) This is may be due to the ability of many of the enterobacteriaceae to have internsic resistance to β-lactams by releasing β-lactamases (Susic, 2004) which break the β-lactam ring and stop the action of antibiotic. SHV-, OXA-,TME-, .CMY-, and CMT-M are the most dominant β- lactamases in Gram-negative bacteria. In our study, blaTEM found in all isolates (100%) that agree with (Medina et al., 2011) who reported that 97.1% of E.coli isolates (226) recovered from lambs affected with diarrhea and from healthy sheep had blaTEM, and (Bailey et al., 2006) who detected blaTEM in all E.coli strains (25) which recovered from faecal flora of healthy humans. Higher than the result of (Park et al., 2005) who detected blaTEM taken from E.cloacae, C.freundii, S.marcescens. also (Hammad and Shimamoto, 2011) who identify blaTEM in 26% of E.coli and K.pneumonia isolates (23) from milk samples. SHV- defined as one of ESBLs which encoded by chromosomal genes in isolates of K. pneumonaie spread by mobile element as plasmids between microbial societies, particularly Enterobacteriaceae family (Paterson and Bonomo, 2005). In this study, SHV-was detected in 80% of the tested isolates which was higher than (Arabi et al., 2013)who reported 53.2% of ESBL –producing isolates have blaSHV. Also, (Tasli and Bahar, 2005) who revealed SHV-in 74.3%. The blaCTX-M is one of the most prevalent ESBLs genes. In this study, blaCTX -gene found 21 isolates of 35 gram negative bacteria (60%). The higher result recorded by (Geser et al., 2012) reported that 78 isolates (85.7%) had CTX- gene fom E.coli, E.cloacae and C.youngae isolates taken from milk samples in Switzerland (Tinelli et al., 2012). In another study carried out by (Valenza et al., 2014) 95.2% of E.coli isolates had CTX-M gene.
Integrons play major role in the prevalence of antibiotic resistance genes among GNB (Rowe-Magnuset al., 2011)Class 1 integrons are spread widely among isolates of family Enteobacteriaceae from animal and human origin. Class1 integrons are the more common and well characterized type (Goldsteinet al., 2001) In our study, class1 integron was detected in 91.4 % of Gram negative isolates, (Figure 5) this result was higher than the result of (Seputeine et al., 2010) who reported that 40% of E.coli that antibiotic resistant load class1 integrons and (Dessie et al., 2013) recorded that E.coli isolated from poultry in Korea carry 54(54.5%) of class 1 integrons. (Wang et al., 2008) identify class1 integron in (56.9%) of E.coli isolated from bovine mastitis. Also (Ahmed and Shimamoto, 2011) found that gram negative bacteria isolated from bovine mastitis in Egypt carry class1 integron. Class 2 integron did not identified in all tested isolates which agreed with the result of (Wang et al., 2008) who could not identify class 2 integron in E.coli from bovine mastitis in Inner Mongolia.While (Ahmed and Shimamoto, 2011) detected class 2 integron in 6 (5.4%) isolates of gram negative bacteria isolated from bovine mastitis in Egypt. Our results revealed a strong relation between multi drug resistance and the high prevelence of class 1 integron.
Conclusion
In Conclusion, mastitis causing economic problem in dairy industry, and the good treatment based on the selection of the suitable antimicrobial drugs. So that, the antibiotic susceptibilities of bacteria recovered from mastitis must be observed periodically. Molecular characterization to this resistance play role in limiting the emergence of this resistance and subsequently decrease the risk of dissemination of resistance bacteria through the food chain.
Acknowledgments
We acknowledged Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Mansoura University, Egypt for support and supply of instruments and devices.
Author’s Contribution
G. Y. designed the experiment and revised the manuscript; A.A performed the tests, analyzed and interpreted the data, and wrote the manuscript. N.A collected samples, isolated strains and participated in manuscript writing. All authors approved the final version of the manuscript for publication.
Conflict of interest
The authors declare that there is no conflict of interest.
References




