Advances in Animal and Veterinary Sciences
Research Article
ESBL Detection and Comparison of Antibiotics Resistance Pattern of Klebsiella Pneumoniae Isolated from Healthy and Acute Respiratory Tract Infected Camels
Sandeep Kumar Sharma1, Kruthika Patel2, Sunil Maherchandani3, Brij Nandan Shringi3
1Department of Veterinary Microbiology and Biotechnology, Post Graduate Institute of Veterinary Education and Research (PGIVER), Jaipur; 2College of Veterinary Medicine, University of Minnesota (USA); 3College of Veterinary and Animal Science, Bikaner Rajasthan University of Veterinary and Animal Sciences (RAJUVAS).
Abstract | Emerging antibiotic resistance is a global problem which interfere with effective disease management in animals. Although antibiotic susceptibility screenings are frequently conducted in human practice, its use in veterinary is very limited and very few literatures are available on antibiotic resistance pattern of Klebsiella pneumoniae isolated from respiratory tract infections of camels. In this investigation, total 65 PCR (16S-23S rDNA internal transcribed spacer region) confirmed K. pneumoniae isolates were included. Among those, 47 were from diseased and 18 were from healthy camels. All isolates were screened for 26 antibiotics of various groups and found 100 % isolates were multidrug resistant, any single isolate from diseased camels was at least resistant to 9 antibiotics and isolate from healthy camel was resistant to more than 6 antibiotics. Out of 26 antibiotics, 100% (65) isolates were susceptible to imepenem but resistant to bacitracin, clindamycin, rifampicin and sulfadiazine. 15 (31.9%) isolates from diseased camels showed Extended Spectrum Beta-Lactamases (ESBL) phenomena while none of the isolate from healthy camels exhibited this activity. Antibiotics susceptibility pattern was found significantly different for seven antibiotics that is cefepime, ciprofloxacin, oxacillin (p≤0.05), erythromycin, norfloxacin, tetracycline and trimethoprim (p≤0.01) whereas non-significant difference for susceptibility pattern was observed for remaining 19 antibiotics. It has established pointer to a correlation between natural and acquired antibiotic resistance and substantial information of occurrence of ESBL activity in K. pneumoniae from camels. Further suggested molecular characterization of these isolates in regards of antibiotic resistance and also the genes responsible for ESBL activity.
Keywords | Klebsiella pneumoniae, ESBL, Antibiotics, Resistance, Camel.
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | January 08, 2017; Accepted | February 09, 2017; Published | February 28, 2017
*Correspondence | Dr. Sandeep Kumar Sharma, Incharge & Assistant Professor, Department of Veterinary Microbiology and Biotechnology Post Graduate Institute of Veterinary Education and Research- Jaipur, Rajasthan University of Veterinary and Animals Sciences, Biknaer- 334001, India; Email: drsharmask01@hotmail.com
Citation | Sharma SK, Patel K, Maherchandani S, Shringi BN (2017). Esbl detection and comparison of antibiotics resistance pattern of Klebsiella pneumoniae isolated from healthy and acute respiratory tract infected camels. Adv. Anim. Vet. Sci. 5(2): 83-91.
DOI | http://dx.doi.org/10.14737/journal.aavs/2017/5.2.83.91
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2017 Sharma et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Klebsiella pneumoniae is ubiquitously present opportunistic pathogen of Enterobacteriaceae family. It is associated with many human and animal ailments that is community acquired pulmonary infections, bone and soft tissue infections, endophthalmitis, endocarditis, bacteremia and a variety of systemic infections in human and metritis in mares, mastitis in cattle, meningitis in piglets and diarrhoea in calves. In addition, K. pneumoniae is an important pulmonary pathogen for camels, horses, dogs, foals, sow and calves (Arora and Kalra, 1973; Carter et al., 1990; Sharma et al., 2013).
Klebsiella pneumoniae is known not only for its virulence but also for its rapidly increasing antibiotic resistance. It expresses significant multidrug resistance by numerous and diverse mechanisms that is one of the best known mechanisms of resistance is the production of antibiotic-inactivating enzymes such as β-lactamases (TEM-1 and SHV-1), carbapenemases (KPC), ampC beta-lactamase and New Delhi metallo-β-lactamase-1 (NDM-1), development of an alternate metabolic pathway and alteration of target sites to prevent the binding of antibiotics, active drug efflux pumps promoting the transport of the antibiotics, impaired uptake of antibiotics due to reduced outer membrane permeability (Pages et al., 2009; Tilak, 2011). Additional mechanisms for quinolone resistance have been described in K. pneumoniae such as mutations in DNA gyrase or in topoisomerase IV, and active efflux of fluoroquinolones (Chevalier et al., 2000).
K. pneumoniae has exhibited natural resistance against some antibiotic groups (Stock and Wiedemann, 2001). But a constant increasing frequency of acquired multidrug resistant K. pneumoniae which possess Extended Spectrum Beta-Lactamases (ESBL) enzymes activity, contributes to unfavorable clinical outcomes, impacts the utilization of resources, increases the burden of effective infection control practice and the overall health economic cost (Marra et al., 2006). Occurrence of antibiotic resistance is huge problem not only in humans but also in animals as antibiotics are mostly used in animals as growth promoters, prophylacticand therapeutic agents (O’Donnell, 2003; Hubert et al., 1991) resulting in excessive use of antibiotics than necessary (Hamud-Socoro et al., 2004). This results in selection pressure and enhances the rapid emergence of multidrug resistance in pathogens of veterinary importance. Many studies are available that define antibiotic resistance pattern of K. pneumoniae from clinical specimens but very little information is available about antibiotic susceptibility pattern or antibiotic resistance among K. pneumoniae as commensal obtained from healthy individuals.
Thus knowledge of multidrug resistance patterns in healthy and diseased individual is essential to enable the selection of appropriate antibiotic therapy and resistance control policies to reduce the spread of antibiotic resistance and mitigate the emergence of new resistant strains of microbes by judicious use of antibiotics (Niederman, 2003). These facts motivate the present study aimed with detection of ESBL activity, occurrence of antibiotic resistance and comparative antibiotic susceptibility pattern of Klebsiella pneumoniae strains obtained from healthy and acute respiratory tract infected (diseased) camels.
Materials and Methods
Bacterial Isolates
In the present investigation, total 65 genotypically confirmed (16S-23S rDNA internal transcribed spacer region based) Klebsiella pneumoniae isolates were examined. Out of these, 47 isolates were obtained from deep nasal swab of clinically acute respiratory tract infected (diseased) camels from clinical complex of college of veterinary and animal science, Bikaner (Rajasthan), India during a wide spread out break of respiratory disease and 18 isolates were obtained from nasal discharge of apparently healthy camels with no history of antibiotic treatment in the last six month.
Antibiotics Susceptibility Testing
Total 26 antibiotics (HiMedia) of various group (Table 1) were examined for antibiotic susceptibility pattern of Klebsiella pneumoniae isolates by disc diffusion method as per technique of Bauer-Kirby (Bauer et al., 1966) by using Mueller-Hinton agar. After inhibition zone measurement, interpretation of resistant, sensitive and intermediates was drown as breakpoints defined by The Clinical & Laboratory Standards Institute (CLSI).
Detection of Extended Spectrum Beta-Lactamase (Esbl) Activity
ESBL activity among Klebsiella pneumoniae isolates was detected by two methods (A) Combined disc method and (B) Double disk test as per techniques described by (Livermore and Brown, 2001; Drieux et al., 2008).
Combined disc method
This method compares the zone of inhibition given by discs containing an extended- spectrum cephalosporin with and without clavulanic acid. An increase of ≥5mm for the discs containing clavulanic acid indicates ESBL production. This method recommends comparison of the zone given by cefotaxime 30µg versus cefotaxime 30 +clavulanic acid 10µg and ceftazidime 30µg versus ceftazidime 30 +clavulanic acid 10µg. The disc types used in present study are available commercially (HiMedia).
Double disk test
This method includes discs containing amoxicillin 20 + clavulanic acid 10 µg (amoxiclav), cefotaxime 30 µg and ceftazidime 30µg. The amoxyclav disc is placed at the centre of the plate. The discs containing cefotaxime and ceftazidime are placed on the either side of the amoxiclav disc at a distance of 25-30mm. The plate is incubated overnight at 37°C, and ESBL production is inferred when the cephalosporin (cefotaxime and ceftazidime) inhibition zone expands towards amoxyclav disc.
Comparative and statistical analysis of antibiotic resistance pattern
For comparison of antibiotic resistance pattern of Klebsiella pneumoniae isolates from infected and apparently healthy animals, the diameter of zone of inhibition for various antibiotics was recorded. Then the mean diameter was calculated for comparison using t- test according to (Campbell et al., 2007).
List of Abbreviations
PCR- Polymerase chain reaction, ESBL - Extended Spectrum Beta-Lactamases, NDM-1- New Delhi metallo-β-lactamase-1.
Results
In the present study all isolates were screened for Extended Spectrum Beta-Lactamases (ESBL) production by both described methods. Out of 47 isolates from acute respiratory tract infected camels, 15 (31.9%) isolates showed ESBL production with varying percentage of resistance towards cephalosporins while none of the isolate obtained from healthy camels showed ESBL activity and resistance towards cephalosporins. All studied isolates were 100% sensitive for imepenem and resistant to bacitracin, clindamycin, rifampicin and sulfadiazine without any significant difference among susceptibility pattern. Additionally, isolates from diseased camels were 100% resistant to amoxicillin, ampicillin, oxacillin and vancomycin in contrast to isolates from healthy camels and for other antibiotics K. pneumoniae isolates showed various percentage of resistance (Table 1). Out of total 65 K. pneumoniae isolates, all isolates were resistant to more than 2 antibiotic while fifty nine (90.76%) isolates were resistant to more than 8 antibiotics. On the basis of multidrug resistance pattern, all isolates were classified into resistotypes thus 31 resistotypes were obtained among isolates from diseased camels and 14 resistotypes were found among isolates from healthy camels (Table 2 and 3). Out of 14 resistotypes from healthy camels none of the resistotype comprised of ciprofloxacin, ceftazidime, norfloxacin, imipenem, gentamicin, kanamycin, ampicillin+ sulbactam, cefepime, cephotaxime and faropenem. In comparison of healthy and diseased camels, for four antibiotics that is erythromycin, norfloxacin, tetracycline and trimethoprim highly significant (p≤0.01)
Table 1: Antibiotic susceptibility of Klebsiella pneumoniae isolates obtained from diseased and healthy camels
|
S. no. |
Name of antibiotic |
Isolates from diseased camels (%) |
Isolates from healthy camels (%) |
||||
|
Sensitive |
Intermediate |
Resistance |
Sensitive |
Intermediate |
Resistance |
||
|
Amoxicillin (AM) |
0 |
0 |
100 |
5.55 |
5.55 |
88.8 |
|
|
1 |
Amoxicillin+ Clavulanic Acid (AC) |
0 |
51 |
49 |
5.55 |
38.8 |
55.5 |
|
2 |
Ampicillin (A) |
0 |
0 |
100 |
0 |
5.55 |
94.4 |
|
3 |
Ampicillin + Sulbactam (AS) |
72.3 |
25.5 |
2.1 |
94.4 |
5.55 |
0 |
|
4 |
Bacitracin (B) |
0 |
0 |
100 |
0 |
0 |
100 |
|
5 |
Cefepime (CPM) |
89.3 |
0 |
10.6 |
100 |
0 |
0 |
|
6 |
Cephotaxime (CE) |
57.4 |
31.9 |
10.6 |
50 |
50 |
0 |
|
7 |
Cephotaxime+Clavulanic Acid (CEC) |
85.1 |
0 |
14.8 |
100 |
0 |
0 |
|
8 |
Ceftazidime (CA) |
70.2 |
36.1 |
12.7 |
94.4 |
5.55 |
0 |
|
9 |
Ceftazidime+ Clavulanic Acid (CAC) |
91.4 |
0 |
8.5 |
100 |
0 |
0 |
|
10 |
Cephalothin (CH) |
53.1 |
23.4 |
23.4 |
55.5 |
44.4 |
0 |
|
11 |
Ciprofloxacin (CF) |
80.5 |
10.6 |
8.5 |
100 |
0 |
0 |
|
12 |
Clindamycin (CD) |
0 |
0 |
100 |
0 |
0 |
100 |
|
13 |
Erythromycin (E) |
0 |
14.8 |
85.1 |
5.55 |
55.5 |
38.8 |
|
14 |
Faropenem (FAR) |
85.1 |
8.5 |
6.3 |
100 |
0 |
0 |
|
15 |
Gentamicin (G) |
95.7 |
4.2 |
0 |
100 |
0 |
0 |
|
16 |
Imipenem (I) |
100 |
0 |
0 |
100 |
0 |
0 |
|
17 |
Kanamycin (K) |
65.9 |
25.5 |
8.4 |
88.8 |
11.1 |
0 |
|
18 |
Norfloxacin (NX) |
82.9 |
8.5 |
8.5 |
100 |
0 |
0 |
|
19 |
Oxacillin (OX) |
0 |
0 |
100 |
5.55 |
11.1 |
83.3 |
|
20 |
Polymyxin-B (PB) |
44.6 |
0 |
55.4 |
55.5 |
0 |
44.4 |
|
21 |
Rifampicin (R) |
0 |
0 |
100 |
0 |
0 |
100 |
|
22 |
Tetracycline (T) |
17 |
65.9 |
17 |
33.3 |
61.1 |
5.55 |
|
23 |
Trimethoprim (TR) |
53.1 |
17 |
29.7 |
100 |
0 |
0 |
|
24 |
Sulfadiazine (SZ) |
0 |
0 |
100 |
0 |
0 |
100 |
|
25 |
Vancomycin (VA) |
0 |
0 |
100 |
0 |
22.2 |
77.7 |
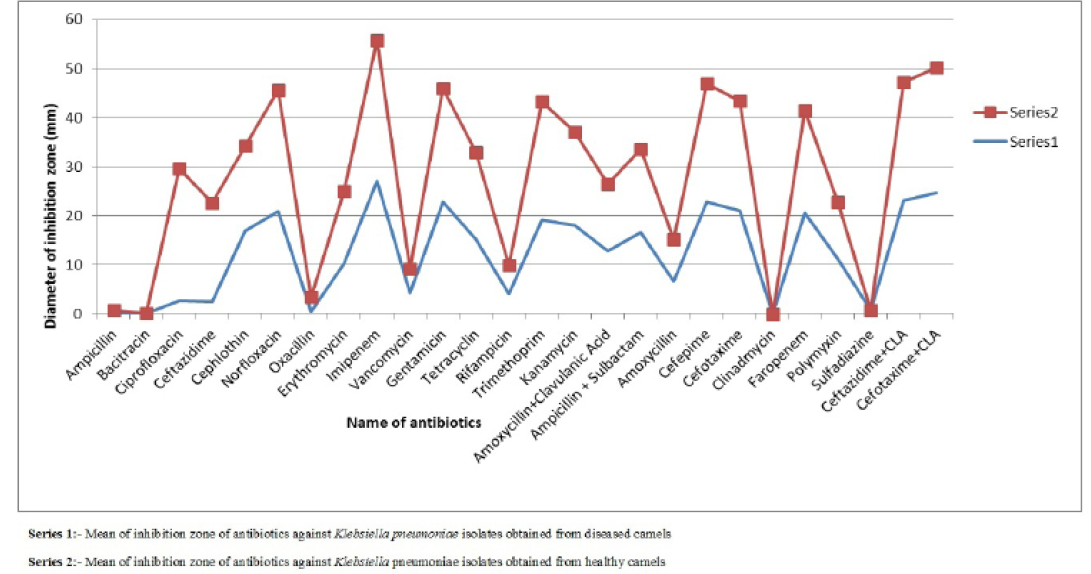
Figure 1: Comparison of mean of diameter of zone of inhibition of antibiotics against Klebsiella pneumoniae isolates obtained from healthy and diseased camels
difference was found in susceptibility pattern of K. pneumoniae isolates and significant (p≤0.05) difference was detected for cefepime, ciprofloxacin and oxacillin (Figure 1 and Table 4) while non-significant differences were observed for resistance pattern of remaining 19 antibiotics among K. pneumoniae strains from healthy and diseased camel (Table 4).
Discussions
Although very few literature is available for Extended Spectrum Beta-Lactamases (ESBL) producing K. pneumoniae from respiratory tract infection of animals. There are studies that reported about 11.1% (three out of 27) ESBL positive strains of K. pneumoniae from intramammary infections of three dairy cow herds (Nobrega et al., 2013; Locatelli et al., 2010) also found 11.11% (one out of nine) ESBL positive K. pneumoniae subsp. pneumoniae from cases of bovine mastitis indicating the occurrence of 3rd generation cephalosporins resistance in veterinary practices. In the present study area, screening of infected animals before treatment is not a common practice resulting in a prolonged and unwise use of antibiotics for treatment being one of the possible reasons for emergence of antibiotic resistance.
It is a well-established fact that new generation antibiotics are more commonly used in human practice than in veterinary, thereby making K. pneumoniae as an important hospital acquired pathogen. The results from this investigation is similar to that of (Damian et al., 2009; Lim et al., 2009) who reported 33.34% and 23.5% ESBL producing strains from human clinical cases respectively. Similarly (Khanfar et al., 2009) reported 17% ESBL positive K. pneumoniae strains from nosocomial and community-acquired infections and all ESBL positive isolates were sensitive to imipenem while in contrast to present study resistant to gentamicin and ciprofloxacin. Findings of present study is also supported by (Gonlugur et al., 2004) with little less frequency (12%) of ESBL producing K. pneumoniae strains from specimen of respiratory tract infected humans and also found that ESBL producing isolates were sensitive to imipenem and ciprofloxacin with more resistance to tetracycline. (Zhou et al., 2011) reported antibiotic resistance pattern and ESBL activity of K. pneumoniae isolated from randomly purchased powdered infant formula and found that 16.6 % isolates were ESBL producers with 100% resistance towards oxacillin and sensitivity for ciprofloxacin and gentamicin, a similar finding in this investigation. (Parasakthi et al., 2000) studied nosocomial outbreak of multidrug-resistant Klebsiella pneumoniae and reported that ESBL producing isolates were completely resistant to ceftazidime and sensitive to imipenem and ciprofloxacin, which is similar to the present study. In the present study, we were able to identify different resistotypes similar to that of (Damian et al., 2009) who classified K. pneumoniae strains in his study into 17 (R1-R16 and S) resistotypes with different susceptibility patterns.The findings of this study are in complete agreement with
Table 2: Multidrug resistance pattern (resistotypes) among Klebsiella pneumoniae isolates from diseased camels
|
Denomination of resistotypes |
Drug resistance pattern |
No. of isolate (%) |
|
PR1 |
A,B,OX,VA,R,AM,CD,SZ,E |
3(6.38%) |
|
PR2 |
A,B,OX,VA,R,AM,CD,SZ,E,AC |
4(8.51%) |
|
PR3 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,PB |
3(6.38%) |
|
PR4 |
A,B,OX,VA,R,AM,CD,SZ,E,PB |
8(17.02%) |
|
PR5 |
A,B,OX,VA,R,AM,CD,SZ,AC,PB |
1(2.12%) |
|
PR6 |
A,B,OX,VA,R,AM,CD,SZ,E,CH |
1(2.12%) |
|
PR7 |
A,B,OX,VA,R,AM,CD,SZ,E,PB,TR |
1(2.12%) |
|
PR8 |
A,B,OX,VA,R,AM,CD,SZ,PB |
2(4.25%) |
|
PR9 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,CH,TR |
1(2.12%) |
|
PR10 |
A,B,OX,VA,R,AM,CD,SZ,E,PB,CH,TR |
1(2.12%) |
|
PR11 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,PB,T |
1(2.12%) |
|
PR12 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,PB,TR |
1(2.12%) |
|
PR13 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,PB,CA |
1(2.12%) |
|
PR14 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,T |
1(2.12%) |
|
PR15 |
A,B,OX,VA,R,AM,CD,SZ,E,PB,TR,CF |
1(2.12%) |
|
PR16 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,CH,CF |
1(2.12%) |
|
PR17 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,TR,CF |
1(2.12%) |
|
PR18 |
A,B,OX,VA,R,AM,CD,SZ,E,TR |
2(4.25%) |
|
PR19 |
A,B,OX,VA,R,AM,CD,SZ,AC |
1(2.12%) |
|
PR20 |
A,B,OX,VA,R,AM,CD,SZ,E,CF |
1(2.12%) |
|
PR21 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,NX |
1(2.12%) |
|
PR22 |
A,B,OX,VA,R,AM,CD,SZ,E,CH,TR,CA |
1(2.12%) |
|
PR23 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,NX,K |
1(2.12%) |
|
PR24 |
A,B,OX,VA,R,AM,CD,SZ,PB,FAH |
1(2.12%) |
|
PR25 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,CH,T,FAH |
1(2.12%) |
|
PR26 |
A,B,OX,VA,R,AM,CD,SZ,E,PB,FAH |
1(2.12%) |
|
PR27 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,PB,CH,TR,T,CPM,CE, |
1(2.12%) |
|
PR28 |
A,B,OX,VA,R,AM,CD,SZ,AC,PB,CH,TR,T,CA,CPM,CE |
1(2.12%) |
|
PR29 |
A,B,OX,VA,R,AM,CD,SZ,AC,CH,TR,T,CA,NX,K,CPM,CE, |
1(2.12%) |
|
PR30 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,PB,CH,TR,T,CA,NX,K,CPM,CE |
1(2.12%) |
|
PR31 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,PB,CH,TR,T,CA,K,AS,CPM,CE |
1(2.12%) |
that of (Mansour et al., 2014) who studied the antibiotic resistance pattern of K. pneumoniae obtained from nasopharyngeal swab of respiratory tract infected camels and other domestic animals. They reported that K. pneumoniae isolates were resistant to β-lactams antibiotics like cefotaxime, ceftazidime, ceftriaxone and aztreonam with 100% resistance to amoxicillin and ampicillin. They also reported that these isolates were sensitive to imipenem, gentamicin, norfloxacin, ciprofloxacin and kanamycin. The antibiotic susceptibility pattern of K. pneumoniae to gentamicin and tetracycline in the present study is in complete agreement with (Al-Doughayam et al., 1999) who reported that isolates of K. pneumoniae from pneumonic camels were 100% sensitive to gentamicin, 16% were sensitive to tetracycline. (El-Enbaawy and Yousif, 2006) also studied K. pneumoniae from sheep pneumonia who reported similar results for ampicillin, norfloxacillin, amoxicillin-clavulanic acid and tetracycline but 100% resistance to gentamicin.
This results of this study agree with (Brosnahan, 2008) who studied antibiogram of clinical specimens of horses including respiratory tract infections caused by K. pneumoniae, reported that imipenem was the only antibiotics to which all isolates were susceptible. Resistance pattern to ampicillin, clindamycin, erythromycin, oxacillin and rifampicin (100% resistant) were also similar. (Sikarwar and Batra, 2011) reported that an isolate of K. pneumoniae from respiratory infections was resistant to all tested antibiotics that is ampicillin, cefotaxime, ceftazidime, tetracycline, ciprofloxacin and gentamicin etc. while other clinical iso
Table 3: Multidrug resistance pattern (resistotypes) among Klebsiella pneumoniae isolates from healthy camels
|
Denomination of resistotype |
Drug resistance pattern |
No. of isolate (%) |
|
HR1 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,PB |
2(11.11%) |
|
HR2 |
A,B,OX,VA,R,AM,CD,SZ |
2 (11.11%) |
|
HR3 |
A,B,OX,VA,R,AM,CD,SZ,AC,PB |
1(5.55%) |
|
HR4 |
A,B,OX,VA,R,AM,CD,SZ,PB |
1(5.55%) |
|
HR5 |
A,B,OX,VA,R,AM,CD,SZ,AC |
3(16.66%) |
|
HR6 |
A,B,OX,VA,R,AM,CD,SZ,E,PB |
1(5.55%) |
|
HR7 |
A,B,OX,VA,R,AM,CD,SZ,E,AC,T |
1(5.55%) |
|
HR8 |
A,B,OX,R,AM,CD,SZ,E |
1(5.55%) |
|
HR9 |
A,B,OX,R,AM,CD,SZ,E,PB |
1(5.55%) |
|
HR10 |
A,B,VA,R,AM,CD,SZ,E |
1(5.55%) |
|
HR11 |
A,B,OX,VA,R,CD,SZ,AC,PB |
1(5.55%) |
|
HR12 |
A,B,OX,R,AM,CD,SZ,AC,PB |
1(5.55%) |
|
HR13 |
A,B,R,AM,CD,SZ,AC |
1(5.55%) |
|
HR14 |
B,VA,R,CD,SZ,CH |
1(5.55%) |
Table 4: Mean± SEM values of diameter of inhibition zone (mm) of antibiotics for Klebsiella pneumoniae isolates obtained from diseased and healthy camels
|
S. No. |
Antibiotic |
Group |
Significance |
|
|
Diseased camels |
Healthy camels |
|||
|
Amoxycillin |
6.57±0.684 |
8.55±1.109 |
Ns |
|
|
1 |
Amoxycillin+ Clavulanic Acid |
12.85±0.560 |
13.64±0.390 |
Ns |
|
2 |
Ampicillin |
0±0.00 |
0.72±0.722 |
Ns |
|
3 |
Ampicillin + Sulbactam |
16.70±0.390 |
16.83±0.389 |
Ns |
|
4 |
Bacitracin |
0.82±0.47 |
0±0 |
Ns |
|
5 |
Cefepime |
22.78±0.584 a |
24.05±0.273b |
* |
|
6 |
Cephotaxime |
21.12±0.761 |
22.33±0.342 |
ns |
|
7 |
Cephotaxime + Clavulanic Acid |
24.70±0.323 |
25.44±0.389 |
Ns |
|
8 |
Ceftazidime |
19.40±0.736 |
20.05±0.481 |
Ns |
|
9 |
Ceftazidime+ Clavulanic Acid |
23.19± 0.319 |
24.05±0.408 |
Ns |
|
10 |
Cephalothin |
16.87±0.860 |
17.27±0.614 |
Ns |
|
11 |
Ciprofloxacin |
24.53±0.89 a |
26.83±0.567 b |
* |
|
12 |
Clinadmycin |
0±0 |
0±0 |
Ns |
|
13 |
Erythromycin |
10.31±0.696 a |
14.72±0.516b |
** |
|
14 |
Faropenem |
20.51±0.399 |
20.94±0.494 |
Ns |
|
15 |
Gentamicin |
22.85±0.478 |
23.05±0.2967 |
Ns |
|
16 |
Imepenem |
27.08±0.645 |
28.66±0.736 |
Ns |
|
17 |
Kanamycin |
18.02±0.542 |
19.05±0.328 |
Ns |
|
18 |
Norfloxacin |
20.97±0.940 a |
24.72±0.774 b |
** |
|
19 |
Oxacillin |
0.46±0.264a |
2.94±1.181 b |
* |
|
20 |
Polymyxin-B |
11.29±0.318 |
11.55±0.293 |
Ns |
|
21 |
Rifampicin |
4.04±0.865 |
5.94±1.055 |
Ns |
|
22 |
Tetracycline |
15.12±0.761 a |
17.88±0.477b |
** |
|
23 |
Trimethoprim |
19.21±1.312 a |
24.05±0.56b |
** |
|
24 |
Sulfadiazine |
0.76±0.535 |
0±0 |
Ns |
|
25 |
Vancomycin |
4.21±0.849 |
5±1.275 |
Ns |
Notes: ‘**’ (p≤0.01) and ‘*’ (p≤0.05) marks significant differences, NS as Non Significat in overall mean values of a parameter among Klebsiella pneumoniae isolates of pneumonic and healthy camels.
lates were susceptible to quinolones and aminoglycosides (over 85%) and chloramphenicol and tetracycline (62%). K. pneumoniae isolated from diarrhea of camel calves were highly resistant to amoxicillin (100%), sulphonamide/trimethoprim (78%) and tetracycline (61%) but sensitive (91.2%) to streptomycin (Glucks, 2007; Seyfried et al., 1989) examined antibiotic susceptibility profiles of Klebsiella pneumoniae strains isolated from different clinical and environmental sources and similar to the present investigation, resistance to aminoglycosides (kanamycin, neomycin, gentamicin, and tobramycin) was found for hospital strains but not for the environmental strains.
In the present study antibiotic resistance was observed in K. pneumoniae isolates obtained from apparently healthy camel that were not treated with antibiotics. This indicates that K. pneumoniae strains have some inherent resistance to antibiotics, as reported by (Stock and Wiedemann, 2001). They found that K. pneumoniae were naturally resistant to amoxicillin, oxacillin, clindamycin, vancomycin, erythromycin, rifampicin and sulfadrugs but sensitive to cephalosporins, carbapenems, aztreonam, quinolones, aminoglycosides and ampicillin/sulbactum similar to our findings in the present investigation. Emergence of resistance for trimethoprim, cephalothin, cephotaxime, cefepime, tetracycline norfloxacin, ciprofloxacin and kanamycin may correlate with the use of these antibiotics during treatment of infected camels.As per the information provided by the veterinarians, these antibiotic were used repeatedly for treating camels at the time of a wide spread outbreak of acute respiratory tract infection in camels in the present study area. It is well doc-umented that widespread and prolonged use of antibiotics prompt to the microbial resistance (Ariffi et al., 2004). In agreement with present study (Loh et al.; 2007) also established the positive correlation between antibiotic intake and microbial resistance who studied antibiotic resistance pattern of K. pneumoniae strains obtained from clinical respiratory specimens of human patients. (Bauernfeind, 1996) reported trend of increasing resistance with time among K. pneumoniae strains for all tested antibiotics except chloramphenicol. They studied antibiotic susceptibility of K. pneumoniae isolates collected in1992 (n= 35) and 1993 (n= 85) from respiratory tract infections of human. The current study provides a pointer to establish a correlation between natural and acquired antibiotic resistance and substantial information of occurrence of ESBL activity in K. pneumoniae obtained from respiratory tract infection of camels. Our next step would be to perform molecular characterization of these isolates to study the transmission of antibiotic resistance and also the genes responsible for ESBL activity in K. pneumoniae isolated from camels.
Conclusions
In the present study, occurrence of ESBL and cephalosporins resistance among isolates from acute respiratory tract infected camels while absence in isolates from healthy camels showed substantial difference in ESBL activity and resistance towards cephalosporins in both source of isolates. Further, more resistotypes were detected among isolates from diseased camels with resistance to more number of antibiotics in comparison of isolates from healthy camels. Statistical analysis indicates highly significant (p≤0.01) and significant (p≤0.05) differences among resistance pattern of isolates from both sources. It may concluded that Klebsiella pneumoniae isolates are continued to evolving antibiotic resistance with significant differences among natural (healthy camels) and acquired (diseased camels) resistance. Further this study suggested genetic explorations of mechanisms of antibiotic resistance to formulate strategy to prevention and control of spread of antibiotic resistance.
Conflict of interest
No conflict of interest exists among authors.
Acknowledgements
We acknowledge the support and facilities provide by Head of department of veterinary microbiology and biotechnology, Dean of college of veterinary and animal science, Bikaner and Dean of Post Graduate Institute of Veterinary Education and Research- Jaipur for this study.
Authors Contribution
Dr. Sandeep Kumar Sharma carried out the conception and design of study, laboratory as well as field work, analysis and writing of the manuscript. Dr. Kruthika Patel contributed in laboratory work, statistical analysis and preparation of the manuscript. Prof. Sunil Maherchandani and Prof. Brij Nandan Shrngi carried out the guidance of conception, preparation of the final version of the manuscript, critical revision of the content. All authors approved the final version of the manuscript for publication.
References





