Advances in Animal and Veterinary Sciences
Research Article
Virulence Properties and Antimicrobial Susceptibility Profiles of Klebsiella Species Recovered from Clinically Diseased Broiler Chicken
Gamal Younis1, Amal Awad1*, Ahmed El-Gamal2, Rania Hosni2
1Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Mansoura University, Mansoura-35516, Egypt; 2Animal Health Research Institute, Mansoura branch, Egypt.
Abstract | E. coli, Klebsiella species are normal intestinal flora of poultry, but could cause infections whenever the immune system of affected bird is compromised. This study was conducted to determine the occurrence of Klebsiella species in clinically diseased chickens with determination of its virulence properties and their antibiotic resistance profile. 200 tissue samples collected from 50 clinically diseased chicken organs (lungs, liver, spleen, and heart) four samples per each bird were screened for the presence of Klebsiella spp. by standard bacteriological methods. Klebsiella spp. isolates were screened for the presence of selected virulence genes including, rmpA (regulator of the mucoid phenotype A), magA (mucoviscosity associated gene) and haemolysin. In addition, Klebsiella spp. isolates were studied for their susceptibility patterns to various antibiotics by disc diffusion method. 30 (15%) isolates were identified as Klebsiella spp. differentiated into 73.33 % (22/30) K. pneumoniae and 26.67 % (8/30) K. oxytoca. Klebsiella isolates were confirmed by PCR using gyrA gene. The rmpA identified in 46.67% (14/30), magA was identified in 53.33% (16/30) isolates, meanwhile, hemolytic activity was detected in 40% (12/30). Klebsiella isolates showed a high resistance to amoxacillin (AX, 100%), amoxicillin /clavulanic acid (100%), piperacillin (86.67%), cefotaxim (86.67%), aztreonam (83.33%), cefapime (70%), ceftriaxone (66.67%) and ciprofloxacin (66.67%), a moderate resistant to chloramphenicol (46.67%), neomycin (33.3%) and norofloxacin (30%). On the other hand, Klebsiella isolates showed the lowest resistance to amikacin (10%). In conclusion, distribution of virulence profile indicates the role of rmpA and magA in pathogenicity of Klebsiella spp. in respiratory infections. Antimicrobial susceptibility pattern showed high multiple antibiotic resistances which require strict regulations antibiotics uses in veterinary therapy.
Keywords | Klebsiella, Virulence genes, PCR, Broiler chicken, Antibiotic resistance
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | October 01, 2016; Accepted | October 10, 2016; Published | October 15, 2016
*Correspondence | Amal Awad, Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Mansoura University, Mansoura-35516, Egypt; Email: amalabdo@mans.edu.eg
Citation | Younis G, Awad A, El-Gamal A, Hosni R (2016). Virulence properties and antimicrobial susceptibility profiles of Klebsiella species recovered from clinically diseased broiler chicken. Adv. Anim. Vet. Sci. 4(10): 536-542.
DOI | http://dx.doi.org/10.14737/journal.aavs/2016/4.10.536.542
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2016 Younis et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Bacterial pathogens play an important role in respiratory disease of domestic poultry species (Glisson, 1998). In many cases, the bacterial pathogens colonize the respiratory system as a secondary bacterial invasion after a primary viral or environmental insult. Klebsiella pneumoniae has been frequently recovered from birds in which it functioned as a primary pathogen and associated with respiratory tract disease (Andra, Jesus and Duarte, 1998). Also, Klebsiella pneumoniae infection of young poultry increased the severity of respiratory disease (Saif et al., 2003).
Klebsiella species are gram-negative, encapsulated, non-motile, rod shape, lactose fermenting bacteria, belong to family Enterobacteriaceae. Members of this family are facultative anaerobic. This genus consist of 77 capsular antigens (K antigens), leading to a different sero-groups (Janda and Abbott, 2006). The organism expresses both O-antigen (smooth lipopolysaccharide) and K-antigen (capsular polysaccharide) and both antigens contribute to its pathogenesis. A major virulence factor of K. pneumoniae is the capsule, which protects Klebsiella from lethal serum factors and phagocytosis (Fung et al., 2002; Mizuta et al., 1983).
The genomic map of K. pneumoniae capsule contains gene clusters as follows: rmpA, rmpA1 and rmpA2 (regulator of the mucoid phenotype A, A1 and A2, respectively), magA (mucoviscosity associated gene A), cps (capsular polysaccharide synthesis), Wb (O-specific polysaccharide is directed by the wb gene cluster) (Regue et al., 2005; Seidler, 1975).
The rmpA and rmpA2 genes regulate the synthesis of the Klebsiella polysaccharide capsule and they are conserved in most isolates of K. pneumoniae. The magA gene is part of the K. pneumoniae serotype K1 capsular polysaccharide gene cluster and contributes to the bacterial virulence (Fang et al., 2004). The magA plays an important role in serious infection of Klebsiella such as septicemia, bacteremia, pneumonia and liver and lung abscesses (Chan et al., 2005; Chung et al., 2007). The chromosomal magA gene causes increased levels of resistance to phagocytosis and has hyperviscous phenotype which is characterized by forming a mucoviscous string during passing loop through a colony (Struve et al., 2005).
In humans, Klebsiella spp. causing infections are often multidrug resistant and an increasing proportion of strains produce extended-spectrum beta-lactamases (ESBLs). Extended-spectrum β-lactamases confer resistance to penicillins and cephalosporins. ESBLs are most commonly detected in K. pneumoniae, they are plasmid-mediated enzymes, and these plasmids also carry resistance genes to other antibiotics. Thus, Gram negative bacilli containing these plasmids are multidrug-resistant (Jacoby, 1997). In contrast, the prevalence of antimicrobial resistance in animal and poultry Klebsiella isolates is poorly documented.
The purpose of this study was to determine some virulence factors associated with Klebsiella spp. infection including, mag A gene (mucoviscosity associated gene) and rmp A (regulator of the mucoid phenotype A), study antimicrobial resistance profile to prevent the spread of resistant K. pneumoniae among diseased chicken via planning a proper control program.
Material and methods
Sampling
This study was conducted in the Department of Bacteriology, Mycology and Immunology at Faculty of Veterinary Medicine, Mansoura University, Mansoura, Egypt. Four samples consisting of heart, liver, lungs and spleen were collected from fifty chickens showed signs of loss of appetite, sneezing, coughing, air sacculitis perhepatitis collected from poultry clinics located in Mansoura city, Egypt giving a total of 200 samples. Samples were collected over a period of 7 months started from February to October, 2015.
Cultural Isolation of the Organism
Bacterial isolation was carried out by inoculating aseptically samples collected from lungs, liver, spleen and heart directly on sheep blood agar and MacConkey’s agar and incubated at 37 °C for 24-48 hrs (Quinn et al., 1994). After incubation, colonies culture characters and morphological characters were studied. Biochemical tests including, catalase, oxidase, indole production, methyl red, Voges-Proskauer, citrate utilization, lysine decarboxylase, urea hydrolysis, lactose fermentation and H2S production were used for Klebsiella spp. identification (Kim et al., 2010; Harada et al., 2013).
Determination of Hemolytic Activity
Klebsiella isolates were tested for the production of B-hemolysin on sheep blood agar plates as described by Gundogan et al. (2011).
Determination of Hypermucoviscosity (HV)
All the obtained Klebsiella isolates were cultivated on sheep blood agar and incubated at 37 °C for 24 hrs. Each bacterial colony was investigated for its hypermucoviscosity. A 5 mm string-like growth was observed and attached to the loop after passing the loop through the colony was considered positive (Zamani et al., 2013).
Extraction of Bacterial DNA
DNA template was prepared from Klebsiella isolates according to Bridge (1996). One ml from distilled water was added to Klebsiella growth on slope agar, then, shacked well. The suspension was centrifuged and the pellet was resuspended again in distilled water by using vortex. To ensure lysis of cells and for complete extraction of DNA, The genomic DNA was extracted by boiling of the suspension for 10 minutes in water bath. The supernatant was used as a template for polymerase chain reaction.
Molecular Identification of of Klebsiella and Virulence Genes
Oligonucleotide primers used in PCR from Metabion (Germany), primers sequence and product shown in Table 1. Preparation of PCR Master Mix according to Emerald Amp GT PCR mastermix (Takara) Code No. RR310A kit: PCR was carried out with Template DNA (6 μl), forward and reverse primers (1 μl), 12.5 μl of Emerald Amp GT PCR mastermix (2 x premix) and 4.5 μl of PCR grade water in a total volume of 25 μl (Aher et al., 2012).
Table 1: Oligonucleotide primers sequences used for amplification of Klebsiella virulence genes
|
arget gene |
Primers sequences |
Amplified segment (bp) |
Reference |
|
gyrA |
CGC GTA CTA TAC GCC ATG AAC GTA |
441 |
Brisse and Verhoef (2001) |
|
ACC GTT GAT CAC TTC GGT CAG G |
|||
|
rmpA |
ACTGGGCTACCTCTGCTTCA |
535 |
Yeh et al., (2007) |
|
CTTGCATGAGCCATCTTTCA |
|||
|
magA |
GGTGCTCTTTACATCATTGC |
1282 |
|
|
GCAATGGCCATTTGCGTTAG |
Temperature and time conditions of the primers during PCR are demonstrated in Table 2 according to specific author and Emerald Amp GT PCR mastermix (Takara) kit. Electrophoresis of amplified products was carried out using 1.5% agarose gel stained with ethidium bromide and detected by UV transillunination. Amplified genes were identified on the basis of fragment size.
Table 2: Cycling conditions of the different primers during PCR
|
Gene |
Denaturation |
Annealing |
Extension |
Final extension |
|
|
1st |
2nd |
||||
|
gyrA |
94˚C 5 min. |
94˚C 30 sec. |
55˚C 45 sec. |
72˚C 45 sec. |
72˚C 10 min. |
|
rmpA |
94˚C 5 min. |
94˚C 30 sec. |
50˚C 45 sec. |
72˚C 45 sec. |
72˚C 10 min. |
|
MagA |
94˚C 5 min. |
94˚C 45 sec. |
50˚C 1 min. |
72˚C 1.2 min. |
72˚C 12 min. |
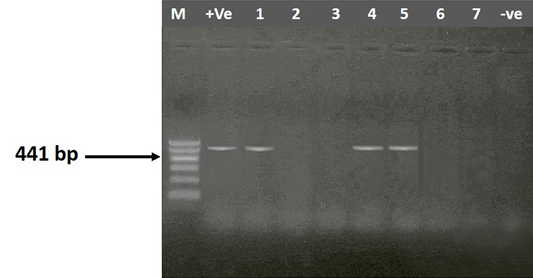
Figure 1: Agarose gel electrophoresis showing amplification of 441bp fragment using gyrA primer. M) 1000 bp DNA Ladder; Lane 1-7) positive samples; +Ve) Positive control; -Ve) Negative control
In vitro Antibiotic Susceptibility Test
The antimicrobial susceptibility profile against Amoxicillin (AX 25), Amoxicillin /clavulanic acid (AMC 20 μg/10 μg), Piperacillin (PRI 100), Cefotaxim (CTX 30), Aztreonam (ATM 30), Cefepime (FEP 30), Ceftriaxone (CRO 30), Ciprofloxacin (CIP 5), Chloramphenicol (C 30), Neomycin (N 30), Norfloxacin (NOR 10), and Amikacin (AK 30) were tested by disk diffusion methods according to Clinical and Laboratory Standards Institute (CLSI formerly NCCLS (CLSI, 2012)).
Results and discussion
Klebsiella species cause infections whenever the immune system of affected bird is compromised (Anonymous, 2006). The isolation of K. pneumoniae and Streptococcus pneumoniae from lungs and trachea could possibly be responsible for the respiratory distress encountered in poultry affected by HPAI during the outbreak (Dashe et al., 2012).
Table 3: Prevalence of Klebsiella species in diseased chicken organs
|
Samples |
No. of samples |
No. (%) of Klebsiella isolates |
||
|
K. pneumoniae |
K. oxytoca. |
Total Klebsiella |
||
|
Lung |
50 |
10 (45.45%) |
4 (50%) |
14 (46.67%) |
|
Liver |
50 |
4 (18.18%) |
2 (25%) |
6 (20%) |
|
Spleen |
50 |
5 (22.72%) |
1 (12.5%) |
6 (20%) |
|
heart |
50 |
3 (13.63%) |
1 (12.5%) |
4 (13.33%) |
|
Total |
200 |
22 (73.33%) |
8 (26.67%) |
30 (15%) |
In the present study, 30 (15%) Klebsiella isolates differentiated into K. pneumoniae 73.33 % (22/30) and K. oxytoca 26.67 % (8/30) were recovered from 200 tissue samples examined conventionally and confirmed by PCR using genus specific primer sequences (gyrA) which yielded product sizes of 441 bp (Figure 1). The isolation rate of Klebsiella spp. was 46.67%, 20%, 20% and 13.33 % from lungs, liver, spleen and heart, respectively (Table 3). The isolation rate of Klebsiella from lungs was higher than the other organs. The wide distribution of K. pneumoniae in the lungs heart, spleen and liver of birds affected could probably indicate concurrent extra–intestinal infections. Türkyilmaz (2005) recorded a higher prevalence of Klebsiella spp., 47.1% from a total of 257 broiler chicken. On the other hand, a low prevalence was recorded by Popy et al. (2011) who isolated Klebsiella spp. from trachea of dead chicken with a percentage of 6% from a total of 50 examined dead chicken. Hinz et al. (1992) isolated Bordetella avium and Chlamydia psittaci as a primary agent of the respiratory disease from dead turkey poults of 6 different flocks and they supposed that by concurrent infections with Klebsiella pneumoniae subsp. pneumoniae, Escherichia coli and Pseudomonas fluorescens causing severe course of the disease. Also, Dashe et al. (2013) isolated K. pneumoniae with a percentage of 8% from lungs and liver of 400 apparently healthy chickens. K. pneumoniae was isolated from birds affected by highly pathogenic avian influenza (H5N1) by Dashe et al. (2008), also he reported that K. pneumoniae was not isolated from H5N1 free flocks which indicated that K. pneumoniae may have acted as a secondary invader to aggravate the clinical signs during H5N1 outbreaks.
In our study hemolytic activity of Klebsiella detected in 40% (12/30). These results consistent with the findings of Albesa et al. (1985), El-Sukhon (2003) and Gundogan and Yakar (2007). Hemolytic activity of Klebsiella spp. was determined with sheep blood agar. Detection of the hemolytic activity of Klebsiella spp. has been reported also by Albesa et al. (1985). The production of hemolysin among gram-negative bacteria is indicative of other virulence and enterotoxigenic factors (Baret and Blake, 1981).
The entire 30 isolates were screened by PCR to identify the mag A and rmp A genes by using specific primer sequences which yielded product sizes of 1282 bp and 535 bp, respectively. Out of the tested isolates, 46.67% (14/30) were positive for rmp A gene (Figure 2) and 53.33% (16/30) were positive for mag A gene (Figure 3). Detection of these genes may indicate the virulence potential of Klebsiella isolates. Capsular serotypes K1 and K2 that carry mag A and rmp A genes make the bacteria more invasive and more resistant to phagocytosis. Prevalence studies showed an association of rmp A with K. pneumoniae virulence (Fang et al., 2004; Ku et al., 2008; Cheng et al., 2010). Yu et al. (2007) reported that there is association between phenotypic evidence of mucoidity and presence of rmpA gene.
The magA gene is the part of capsular polysaccharide gene of K. pneumoniae serotype k1 (Fang et al., 2005). Struve et al. (2005) described mag A as a novel virulence factor responsible for the increased virulence of certain K. pneumoniae strains. They provide evidence that the mag A gene, so far believed to be a specific virulence factor in highly virulent Klebsiella strains.
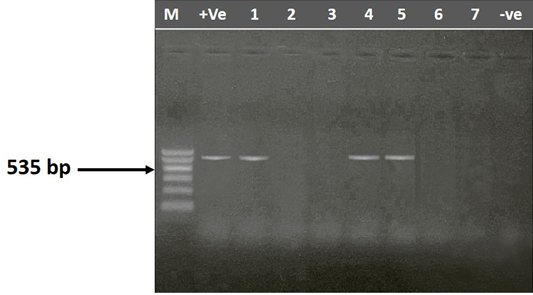
Figure 2: Agarose gel electrophoresis showing amplification of 535 bp fragment using rmpA primer. M) 1000 pb DNA ladder; Lane 1, 4 and 5) Positive samples; Lane 1, 2, 6 and 7) negative samples; +Ve) Positive control; -Ve) Negative control
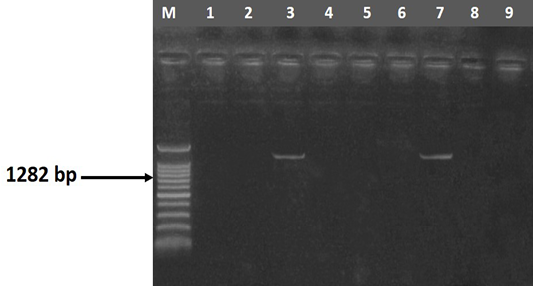
Figure 3 Agarose gel electrophoresis showing amplification of 1282 bp fragment using magA primer. M) DNA ladder; Lane 3 and 7) Positive samples; Lane 1, 2, 5, 6, 8 and 9) Negative samples
Table 4: Percentages of Antimicrobial resistance of Klebsiella isolates from diseased chicken birds
|
Sensitive |
Resistant |
Antimicrobial class |
Disc Code |
Antibiotic |
|
0 (0.00%) |
30 (100%) |
ß-Lactams |
AX |
Amoxacillin |
|
0 (0.00%) |
30(100%) |
ß-Lactam/ ß-lactamase inhibitors |
AMC |
Amoxicillin/ clavulanic acid |
|
4 (13.33%) |
26 (86.67%) |
ß-Lactams |
PRI |
Piperacillin |
|
4 (13.33%) |
( 86.67%) 26 |
Cephalosporins |
CTX |
Cefotaxim |
|
5(16.67%) |
25(83.33%) |
Monobactams |
ATM |
Aztreonam |
|
9 (30%) |
21 (70%) |
Cephalosporins |
FEP |
Cefepime |
|
10(3.33%) |
20 (66.67%) |
Cephalosporins |
CRO |
Ceftriaxone |
|
10 (33.33%) |
20(66.67%) |
Quinolones |
CIP |
Ciprofloxacin |
|
16 (53.33%) |
14 (46.67%) |
Phenicols |
C |
Chloramphenicol |
|
20 (66.67%) |
10 (33.33%) |
Aminoglycosides |
N |
Neomycin |
|
21 (70%) |
9 (30%) |
Quinolones |
NOR |
Norofloxacin |
|
27 (90%) |
3(10%) |
Aminoglycosides |
AK |
Amikacin |
In this study, hyperviscosity (HV) was detected in 16 out 30 Klebsiella isolates. HV phenotype was highly correlated with the presence of the rmpA gene. Of 16 mucoid isolates, 81.25% (13/16) were rmpA gene positive, and 18.75% (3/16) were rmpA gene negative. Among non-mucoid (14) isolates, 92.86% (13/14) were negative for the rmpA gene and 7.14% (1/14) were rmpA positive. In a previous study, Yu et al. (2006) showed that prevalence of magA, rmpA and HV were 17%, 48% and 38%, respectively. Amraie et al. (2014) proved that out of 173 isolates of K. pneumoniae, 42.19% and 2.31 % of samples were positive for HV and magA, respectively.
Klebsiella isolates were tested against 12 antimicrobial agents using the Kirby-Bauer disk diffusion assay. All of the isolates were resistant to amoxicillin (AX). AX resistance has been reported in veterinary clinical isolates (Brisse and Van Duijkeren, 2005). Most Klebsiella isolates are naturally resistant to ampicillin due to a constitutively expressed chromosomal class Ab-lactamase (Livermore, 1995).
Different rates of cephalosporin resistance had been reported. In this study, the resistance rates for cephalosporin were 86.67% for cefotaxime, 70% for cefapime 66.67% for ceftriaxone which was not far from those previously reported by Singh and Goyal (2003). On the other hand, a lower rate of resistant to cephalosporins was recorded by Ullah et al. (2009).
Resistance of Klebsiella isolates to ciprofloxacin in our study was 66.67% (Ullahet al., 2009). Villegas et al. (2004) reported a lower level of resistance of Klebsiella to ciprofloxacin. Ciprofloxacin is a broad spectrum fluoroquinolone antibacterial agent (Periti et al., 1998) the resistance of Klebsiella to ciprofloxacin mainly due to a chromosomal mutation in the gyrA gene, which codes for the target of quinolone activity (Bagel et al., 1999).
In concerning to aztreonam, the rate of resistance to aztreonam was 83.33%. It is a synthetic monocyclic b-lactam in the family of monobactams and is exclusively active against the aerobic gram-negative bacilli (Sader et al., 2003). According to Nijssen et al. (2004), aztreonam had moderate activity against K. pneumonia and K. oxytoca.
Aminoglycosides are active against gram-negative bacilli which have a clinical important (Ramirez and Tolmasky, 2010). In this study, Klebsiella isolates showing 10% resistance to amikacin. A low prevalence of amikacin resistance (7%) to Klebsiella spp. isolated from meat samples were also reported by Gundogan et al. (2011). On the other hand, Ullah et al. (2009) reported that 63.04% of Klebsiella isolates were susceptible to amikacin.
Klebsiella strains recorded high antibiotic resistance with multiple antibiotic resistance (MAR). Rate of multiple antibiotic resistances was extremely high which may be due to the hazard routinely use antibiotics for treatment and control of bacterial diseases in poultry farms. When these antibiotics are administered to the birds at low levels for a long period, certain bacterial species become resistant (Kilonzo-Nthenge et al., 2008). These antibiotic-resistant bacteria can reach to human through consumption of food products from animal origin and by direct contact (Van den Bogaard and Stobberingh, 2000). Our finding is in accordance with a previous literature reported by Davies et al. (2016) who reported a 25% multiple drug resistance of K. pneumoniae isolates from passerin and psittacines. Bonnedahl et al. (2014) detected 13% MDR strains from the samples they collected from ageese and free-living gulls in Alaska, USA.
Conclusion
This study highlights the prevalence of virulence attributes of klebsiella spp. in clinically diseased chicken suffering from respiratory manifestation at Mansoura city, Egypt. The presence of these virulence genes confirmed the pathogenic potential of the isolated strains and their association with clinical manifestations in respiratory tract infections of broiler chicken. Antimicrobial susceptibility pattern showed high multiple antibiotic resistances which require strict regulations of the use of antibiotics in veterinary therapy to minimize the emergence of resistant bacteria in animals which may increase the public health problem.
Acknowledgments
Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Mansoura University, Egypt, supported this study.
Author’s Contribution
Gamal Younis designed the experiment and revised the manuscript. Amal Awad wrote the paper and shared in carry out the practical part and took the responsibility of correspondence to the journal. Ahmed El-Gamal shared in the collection of samples. Rania Hosni collected chicken samples and carried out the practical part. All authors approved the final version of the manuscript for publication.
Conflict of interest
The authors declare that there is no conflict of interest.
References





