Advances in Animal and Veterinary Sciences
Research Article
Genetic Diversity and Phylogenetic Relationship among three Duck Breeds and Geese using RAPD Markers
Heba Abdou Basha1*, Walaa Slouma Hamouda Abd el Naby1, Hanim Shabaan Mohammed Heikal2
1Department of Animal Husbandry and Animal Wealth Development, Faculty of Veterinary Medicine, Alexandria University, Egypt; 2Department of Animal Husbandry and Animal Wealth Development, Faculty of Veterinary Medicine, Sadat City University, Egypt.
Abstract | Poultry and poultry products are the most economical protein sources for the ever-increasing human population and to meet the future food security. While these are promising food sources, the importance of genetically improved avian species in the breeding programme has increased significantly. In this study, we applied randomly amplified polymorphic DNA (RAPD) methodology to elucidate genetic variations and degree of relatedness among native Egyptian breed of geese and ducks (Muscovy, Sudani and White Pekin). In this regard, 19 random primers were designed and screened and a total of 16 primers produced a total of 189 reproducible amplified fragments. The 169 DNA bands were polymorphic with percentage presentation of 89.41, however, OPA04 and OPA03 primers have generated high polymorphic bands. Several primers have produced bands, which were specific for each tested species, suggesting their potential to be used as a species- or breed-marker. Phylogenetic tree, based on the linkage distance, revealed close relationships among Geese, Muscovy and Sudani duck breeds, however, White Pekin showed a clear distance from rest of the studied breeds. Taken together, presented results provide foundation studies for the selection of breeds under limited resource settings.
Keywords | RAPD-PCR, Geese, Ducks, Polymorphism
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | July 17, 2016; Accepted | August 24, 2016; Published | September 12, 2016
*Correspondence | Heba A Basha, Department of Animal Husbandry and Animal Wealth Development, Faculty of Veterinary Medicine, Alexandria University, Egypt; Email: henour_hnour@yahoo.com
Citation | Basha HA, Abd el Naby WSH, Heikal HS (2016). Genetic diversity and phylogenetic relationship among three duck breeds and geese using RAPD markers. Adv. Anim. Vet. Sci. 4(9): 462-467.
DOI | http://dx.doi.org/10.14737/journal.aavs/2016/4.9.462.467
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2016 Basha et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
With the increasing human population, food security has emerged as an important issue for the sustainability of the mankind. It is estimated that by 2020, the consumption of the current food rate would increase by 20%. This places a heavy burden on existing food resources including poultry and poultry products. After chicken, ducks are considered the most important and common component of the poultry industry, around the world. Duck production has received immense attention due to its higher profitability compared to other poultry species (El-Soukkary et al., 2005), mainly due to higher feed conversion ratios. Ducks can utilize cheap raw material and produce significant amount of palatable meat and large number of eggs in a short period. Additionally, ducks and geese had a historic importance in Egypt as they found on the walls of ancient temples. Sudani breed is considered a local breed that has adapted to the Egyptian environmental conditions and has gained heat tolerant compared to White Pekin and Muscovy duck which are regarded as exotic breeds.
White Pekin ducks originated from mallard duck and are reared mainly for meat production due to their high growth rate. It has a clean dressed carcass, with subcutaneous fatty layer. While Muscovy duck seems an odd duck, it is the only domestic duck which is not originated from wild Mallard. It has some common morphological and productive characters to mallard and resembles geese for other characters. Body morphology of Muscovy duck resembles to Mallard compared to Geese. Muscovy ducks have abilities to graze and have 35 days of incubation period similar to Geese. Thus, technically Muscovy ducks are considered geese however genetically these are not yet categorized.
To improve ducks productivity, genetic information as well as genetic variation among different breeds has to be identified. With recent development of molecular techniques, it became easier to improve genetics of the livestock. Randomly amplified polymorphic DNA (RAPD) is widely used for studying genetic variations in populations, identification of species and breed of specific species (William et al., 1990; Smith et al., 1996; Yoon and Kim, 2004).
The RAPD is based on amplification of genomic DNA by arbitrator short primers. In previous studies, RAPD has been applied to investigate genetic similarity, diversity and relationship among different avian species such as chicken (Baloza et al., 2014), turkey (Smith et al., 1996), geese (Maciuszonek et al., 2005; Devrim et al., 2007) and ducks (Su et al., 2006). The objective of the present study is to evaluate the genetic variations among three duck breeds and their genetic relationships with geese. Results would provide foundation for the selection of required traits and phenotypes that facilitate the improvement of duck productivity.
Material and Methods
Blood Samples
Five mature birds of local and native Egyptian geese, and three duck breeds [including two exotic breeds, Muscovy (M) and White pekin (WP), and one local breed, Sudani (S)] were used in this study. Blood samples were collected from the wing vein in sterilized vacuum tubes containing ethylenediamine tetra acetic acid (EDTA) as anticoagulant and were placed directly on ice after collection. These samples were then stored at -20 ºC until further used.
Extraction of DNA, RAPD PCR Amplification and Gel Electrophoresis
Genomic DNA was extracted from blood samples of each bird using DNA Extraction kit (Thermo Scientific, Germany) according to the manufacture instruction. The quality of isolated DNA was assessed through 1.5% agarose gel electrophoresis, and DNA of good quality with intact bands and without smearing were visualized. Pooling of five DNA samples in each breed was prepared by mixing equal amount of DNA for screening of random primers.
A total of 19 random 10-mer primers (Table 1) were used to detect the polymorphism among geese and three duck breeds: Muscovy, White Pekin and Sudani. The amplification of the PCR products was performed in a total of 25 µl of reaction using the procedure described by Williams et al. (1990) with some modification. The PCR reaction mixture consists of 2 µl genomic DNA, 2.5 µl of 10x Dream Taq Green buffer (Thermo Scientific, Germany), 5 μl primer (10 pmole), 0.5 μl of dNTPs(10 mM) (Thermo Scientific, Germany), 0.3 μl of Dream Taq DNA polymerase (Thermo Scientific, Germany) and 14.7 dH2O. Amplification was carried out in the thermal cycler (Techno, UK) using following cycling profile: initial denaturation at 95°C for 5 min, followed by 40 cycles at 95°C for 1 min, annealing temperature as shown in Table 1 for 1 min and extension at 72°C for 1 min, final extension step at 72°C for 10 min.
PCR products (10 μl) and 5 μl (100 bp) DNA ladder (Thermo Scientific, Germany) were electrophoretically separated in 3% agarose gel stained with ethidium bromide. Electrophoresis was carried out for 60 min at 120 volts, then electrophoresis gel was examined in the UV transilluminator and bands were visualized and photographed using gel documentation system (InGenius, Syngene Bio Imaging, USA).
Table 1: Name and sequence of primers used in RAPD-PCR assay
|
Primer |
Sequence (5'-3') |
Annealing temperature |
Primer |
Sequence (5'-3') |
Annealing temperature |
|
OPA03 |
AGTCAGCCAC |
32°C |
CH 1 |
GAATGCGACG |
34°C |
|
OPA04 |
AATCGGGCTG |
32°C |
CH 2 |
ATGACGTTGG |
34°C |
|
OPA05 |
AGGGGTCTTG |
32°C |
CH 3 |
CTGAGGAGTG |
34°C |
|
OPA07 |
GAAACGGGTG |
32°C |
CH4 |
GGGCTAGGGT |
34°C |
|
OPA08 |
GTGACGTAGG |
32°C |
CH 5 |
ACCGGGAACG |
34°C |
|
OPA10 |
GTGATCGTAGG |
32°C |
-132 |
AGCGATCTCC |
32°C |
|
OPA12 |
TCGGCGATAG |
32°C |
-115 |
TTCCGCGGGC |
32°C |
|
OPA17 |
GACCGCTTGT |
34°C |
-127 |
ATCTGGCAGC |
32°C |
|
OPA19 |
CAAACGTCGG |
34°C |
-134 |
AACACACGAG |
34°C |
|
-137 |
GGTCTCTCCC |
34°C |
Table 2: Total number of bands, number and percentage of polymorphic bands, number of common bands and number of species specific bands generated by sixteen primers in the geese and three duck breeds
|
Primer |
Total no. of bands |
No. of polymorphic bands |
% of polymorphic bands |
No. of common bands |
No. of species specific bands |
Molecular size ange(bp) |
||||
|
G |
M |
S |
WP |
Max. |
Min. |
|||||
|
OPA03 |
21 |
20 |
95.23 % |
1 |
- |
- |
5 |
1 |
1400 |
260 |
|
OPA04 |
27 |
24 |
88.88 % |
3 |
5 |
2 |
1 |
3 |
1250 |
160 |
|
OPA05 |
13 |
11 |
84.61 % |
2 |
5 |
- |
2 |
1 |
1490 |
510 |
|
OPA08 |
10 |
9 |
90.00 % |
1 |
1 |
4 |
- |
1 |
900 |
310 |
|
OPA10 |
18 |
13 |
72.22 % |
5 |
2 |
- |
- |
3 |
1490 |
290 |
|
OPA12 |
3 |
3 |
100. % |
- |
- |
- |
1 |
- |
1250 |
250 |
|
OPA17 |
7 |
7 |
100 % |
- |
1 |
4 |
1 |
1 |
1000 |
320 |
|
OPA19 |
3 |
3 |
100 % |
- |
- |
- |
- |
2 |
1400 |
390 |
|
CH3 |
12 |
11 |
91.66 % |
1 |
3 |
1 |
- |
1 |
Above 1500 |
390 |
|
CH4 |
12 |
9 |
75.00 % |
3 |
- |
- |
- |
- |
1500 |
310 |
|
CH5 |
11 |
11 |
100 % |
- |
2 |
- |
2 |
- |
Above 1500 |
430 |
|
-115 |
8 |
8 |
100 % |
- |
- |
3 |
- |
- |
1250 |
320 |
|
-127 |
7 |
7 |
100 % |
- |
- |
- |
- |
3 |
700 |
490 |
|
-132 |
15 |
15 |
100 % |
- |
2 |
1 |
- |
4 |
Above 1500 |
260 |
|
-134 |
9 |
7 |
77.77 % |
2 |
- |
- |
- |
- |
Above 1500 |
480 |
|
-137 |
13 |
11 |
84.61 % |
2 |
3 |
- |
- |
2 |
1500 |
320 |
|
Total |
189 |
169 |
89.41 % |
20 |
24 |
15 |
12 |
22 |
- |
- |
Statistical Analysis and Phylogenetic Tree Construction
The PCR products were scored across the lanes, the presence or absence of distinct bands were recorded as (1) and (0) respectively, in RAPD profile of geese and three duck breeds. Patterns between different breeds were compared using the similarity index. This index reflects the extents of band sharing, and calculated using the formula (Lynch, 1990):
BS = 2Nab / (Na+ Nb)
Where;
Nab are the number of bands shared by breed a and b.
Na and Nb are the total number of fragments scored in breed a and b, respectively.
BS values were calculated for each primer separately and the average for all primers was carried out with each comparison.
According to Sneath and Sokal (1973), dendrogram was constructed for estimation genetic similarity. The similarity matrix was analyzed by the unweighted pair group methods with arithmetic average (UPGMA). The cluster analysis and dendrogram construction was performed with Statistica 5 (1995).
RESULTS
Banding Pattern
The banding profiles generated through RAPD assay in the present study were considered to differentiate between geese and three duck breeds in Egypt to clarify genetic diversity and relatedness. Sixteen primers out of nineteen have generated total of 189 positive and detectable DNA bands. The score varied from 3 bands, produced by OPA12 and OPA19, to 27 DNA bands produced by OPA04 with approximately size ranged from 160bp to above 1500bp.
From a total of 189 DNA bands, 169 DNA bands (89.41%) were polymorphic (Table 2). The percentage of polymorphism produced by each primer differed from one primer to another. It was ranged from 72.22% produced by primer OPA10 to 100% produced by OPA12, OPA17, OPA19, CH5, -127, -115 and -132. Seventy three DNA fragment bands were expressed as species specific bands, where 24 species specific bands were detected in Geese, 15, 12, 22 species specific bands were detected in Muscovy, Sudani and White pekin ducks, respectively (Table 2).
Primer OPA12 at approximately molecular weight of 250 bp produced only one unique band in Sudani breed (Figure 1). Thus, this primer can be used for characterization of this duck breed. Additionally, primer OPA19 and primer -127 generated only two DNA bands (390 bp and 1400 bp) and three monomorphic bands (500, 550 and 700bp) in White Pekin breed (Figure 1). While, Primer -115 gave three unique DNA bands (Table 2) with approximately molecular weight 320, 400 and 425bp for Muscovy breed (Figure 2).
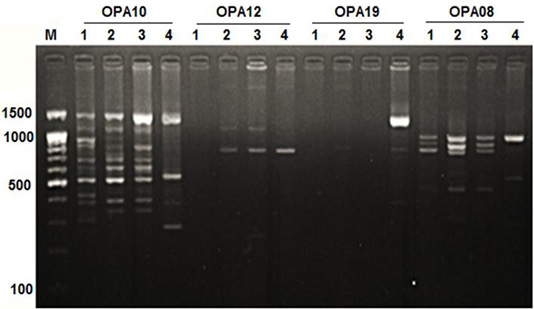
Figure 1: RAPD PCR patterns generated by primer OPA10, OPA12, OPA19 and OPA08. The fragment patterns are identified as; Lanes 1: Geese; Lanes 2: Muscovy; Lanes 3: Sudani; Lanes 4: White Pekin; Lane M: DNA marker (100 bp DNA ladder was loaded)
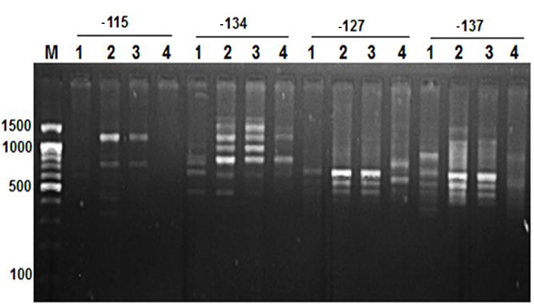
Figure 2: RAPD PCR patterns generated by primer -115, -134, -127 and -137. The fragment patterns are identified as; Lanes 1: Geese; Lanes 2: Muscovy; Lanes 3: Sudani; Lanes 4: White Pekin; Lane M: DNA marker (100 bp DNA ladder was loaded)
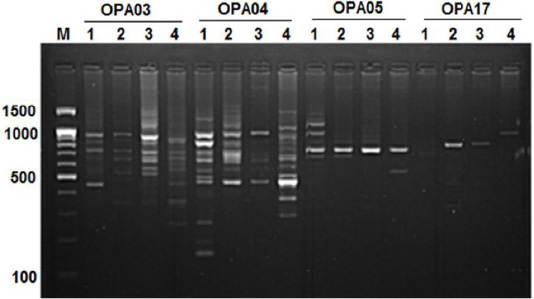
Figure 3:RAPD PCR patterns generated by primer OPA03, OPA04, OPA05 and OPA17. The fragment patterns are identified as; Lanes 1: Geese; Lanes 2: Muscovy; Lanes 3: Sudani; Lanes 4: White Pekin; Lane M: DNA marker (100 bp DNA ladder was loaded)
DNA fragment bands profile with primer OPA17 produced seven DNA bands, which were specified for Geese, Sudani and White pekin ducks and four DNA bands were specified for Muscovy ones (Figure 1).
The highest number of DNA fragments produced by primer OPA04 with polymorphic 88.88%. Results revealed the highest species-specific bands (11 DNA bands) among studied samples were generated by OPA04 primer with approximately molecular weight ranged from 160bp to 1250 bp (Figure 3). Here five monomorphic bands were detected in geese and 2, 1, 3 species specific bands showed for Muscovy, White Pekin and Sudani breed, respectively.
In geese, OPA05 primer revealed five monomorphic bands with approximate molecular weights of 600, 700, 1100, 1300 and 1490 bp (Figure 3). But for duck breeds, this primer produced two unique bands (1000 bp and 1400 bp) in Sudani breed and one unique band 510 bp in Whit Pekin breed. In addition, primers OPA12 and OPA19 didn’t show any band in geese (Figure 1).
On the other hand, primer CH4 and primer -134 generated nine and seven polymorphic DNA bands (Figure 2 and 4) in studied geese and duck breeds (Table 2) without any species-specific bands. Therefore, they revealed the highest similarity results among such species.
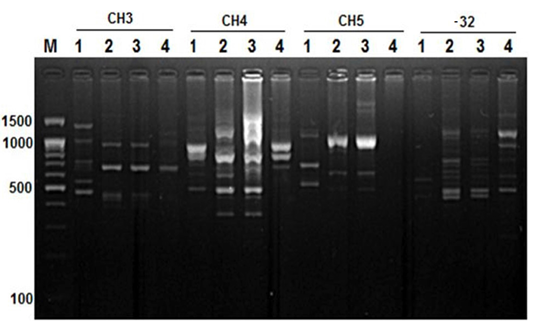
Figure 4: RAPD PCR patterns generated by primer CH3, CH4, CH5 and primer -32. The fragment patterns are identified as; Lanes 1: Geese; Lanes 2: Muscovy; Lanes 3: Sudani; Lanes 4: White Pekin; Lane M: DNA marker (100 bp DNA ladder was loaded)
Table 3: Similarity coefficients among geese and three duck breeds based on RAPD data
|
Species |
Similarity coefficients |
|
Muscovy ducks and geese |
0.900 |
|
Sudani ducks and geese |
0.903 |
|
White Pekin ducks and geese |
0.898 |
|
Muscovy and Sudani ducks |
0.934 |
|
Sudani and White Pekin ducks |
0.899 |
|
Muscovy and White Pekin ducks |
0.894 |
Similarity and Phylogenetic Relationships
The highest similarity value was found between Muscovy and Sudani ducks (Table 3), while the lowest was appeared between Muscovy and White Pekin ducks (0.894).
Based on genetic distance values, the phylogenetic tree of studied geese and the three duck breeds was constructed. The dendogram (Figure 5) describe the genetic relationship among three duck breeds (Muscovy, Sudani and White Pekin) and Geese. Muscovy and Sudani breeds appeared to be the most closely related breeds and geese showed close similarity to them. However, White Pekin was clustered distinctly and separately from the rest of studied breeds.
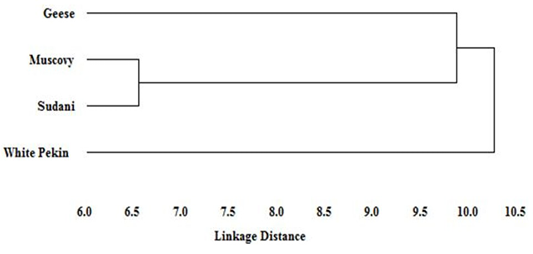
Figure 5: Dendogram of the phylogenetic relationships among Geese and Muscovy, Sudani and White Pekin duck breeds based on genetic distance
Discussion
RAPD technique has been used in many studies to assess genetic relationships within waterfowls. However, it remains to be assessed if RAPD can be applied to study the genetic diversity among duck breeds and geese.
In the present study the RAPD technique was used to assess genetic relationships among Egyptian native geese, two exotic duck breeds (Muscovy and White Pekin) and one local breed (Sudani). Sixteen primers from nineteen primers revealed 169 polymorphic bands (89.41%). The percentage of polymorphism produced by each primer varied from 72.22% to 100% among studied breeds. Previous studies showed different percentages of polymorphic bands using RAPD analysis in duck and geese, as 83% in five ducks closed population in Egypt (El-Gendy et al., 2005), 66.78% in Chinese ducks (Su et al., 2006), 70% in Indian native ducks (Sankhyan, 2007), 69.8% in Moti native ducks (Alyethodi et al., 2010) and 83.3% in local geese with four different feather colors (Devrim et al., 2007).
From using primers, few primers showed unique band for each specie as in OPA05 revealed two unique bands in Sudani breed (1000 bp and 1400 bp) and one unique band in Whit Pekin breed (510 bp). Also, Primer -115 produced three unique DNA bands (Table 2) for Muscovy breed and Primer OPA12 generated only one unique band for Sudani breed. Moreover, the highest species specific bands among studied samples were generated by OPA04 primer; five monomorphic bands were detected in Geese and 2, 1, 3 species specific bands showed for Muscovy, Sudani and White Pekin breeds. Therefore, we suggest that this primer can be used as species-specific markers. Similarly, Maciuszonek et al. (2005) obtained from 1 or 2 up to 13 specific bands per geese line (Kartuska, Labelaska, Kielecka and Padkarpacka) and suggested their potential for use as population specific markers. In addition, Sharma et al. (2001) showed the presence of monomorphic band specific for different chicken breeds.
Little information is available about the origin of Sudani duck. Thus an evolutionary dendogram relationship was constructed based on linkage distance. The dendogram showed that Sudani and Muscovy breeds shared the same cluster as has previously been reported with Abd-Elmaksoud et al. (2009) and Abd el Samee et al. (2012) who suggested that Sudani duck originated from the same ancestor of Muscovy duck (Carina moschata). However, White Pekin duck had clustered apart from other studied breeds. These results showed a similar trend as El-Gendy et al. (2005) who reported similar dendogram pattern among different duck breeds.
Moreover, distance phylogenetic relationship revealed that geese are closer to Muscovy duck than Sudani and Pekin breeds. This result, clarify the known common productive traits between Muscovy duck and geese, Muscovy is the only duck breed which incubate their eggs for 35 days as Chinese and Egyptian geese, which suggest that Muscovy duck and geese may share similar genetic composition.
Gholizadeh et al. (2007) have used the RAPD technique to evaluate the genetic diversity among Muscovy, Pekin, Native and Khaki Campbell duck breeds. They showed that the genetic similarity between Muscovy and Pekin breeds was 0.73, whereas, genetic similarity between the pervious breeds in the present study was 0.895. However, the genetic similarity was higher between Muscovy and Sudani ducks compared to Muscovy and White Pekin breeds.
Conclusion
The RAPD is a useful technique for detecting the polymorphism and genetic relationship among geese and different duck breeds. The genetic diversity was the lowest between Muscovy and Sudani breeds, and the highest between White Pekin breed and the other duck studied breeds, which could be exploited for improving productivity by selective breeding. The genetic distance between Muscovy and geese was the shortest indicating the genetic similarity between these two species. Further investigations are required to assess other traits with these selected breeds to obtain full potential of productivity and to meet the increasing demand of food in expanding human population.
ACKNOWLEDGMENTs
We would like to show our gratitude to our professors and colleagues who provided expertise that greatly assisted the research.
Conflict of interest
There is no conflict of interest.
Authors’ contribution
All authors contributed equally to carry out this work.
REFERENCEs





