Advances in Animal and Veterinary Sciences
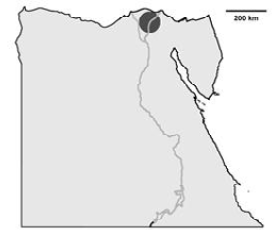
The location of the collected IBV samples in Egypt
Schematic illustration of Egypt and the area where the IBV isolates have been detected (dark gray). The IBV isolates are distributed in El-Dakahlyia and Damietta provinces in the Nile delta area of northern Egypt.
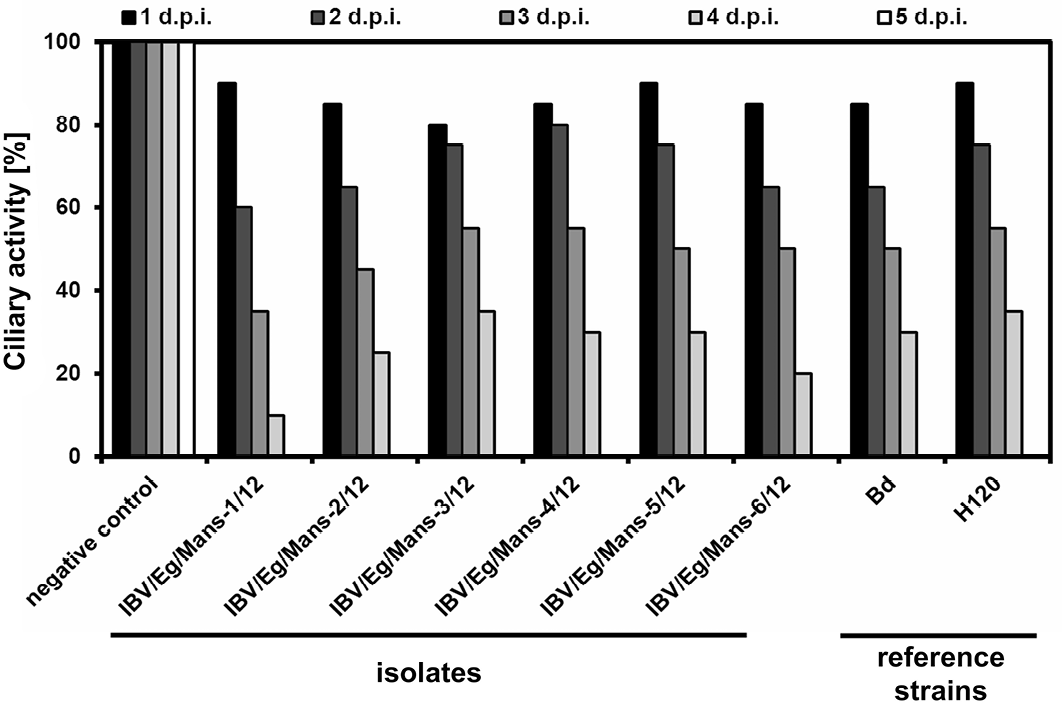
Ciliostatic analyses of TOCs inoculated by the IBV isolates and reference strains
Chicken TOCs inoculated by 1x104 ffu/ml of the IBV isolates (IBV/Eg/Mans-1, -2, -3, -4, -5, or -6/12) and the reference strains (Beaudette and H120); Mock-treated TOCs served as a negative control
Sequence identities on the nucleotide and amino acid level of the Egyptian IBV isolates to reference strains based on their partial S1-protein sequences
By RT-PCR, partial S-protein sequences (A) were determined and used to calculate the total sequence identity on nucleotide (B) and amino acid level (C) for the Egyptian IBV isolates to selected reference strains . Numbers indicate the total sequence identity (in %).
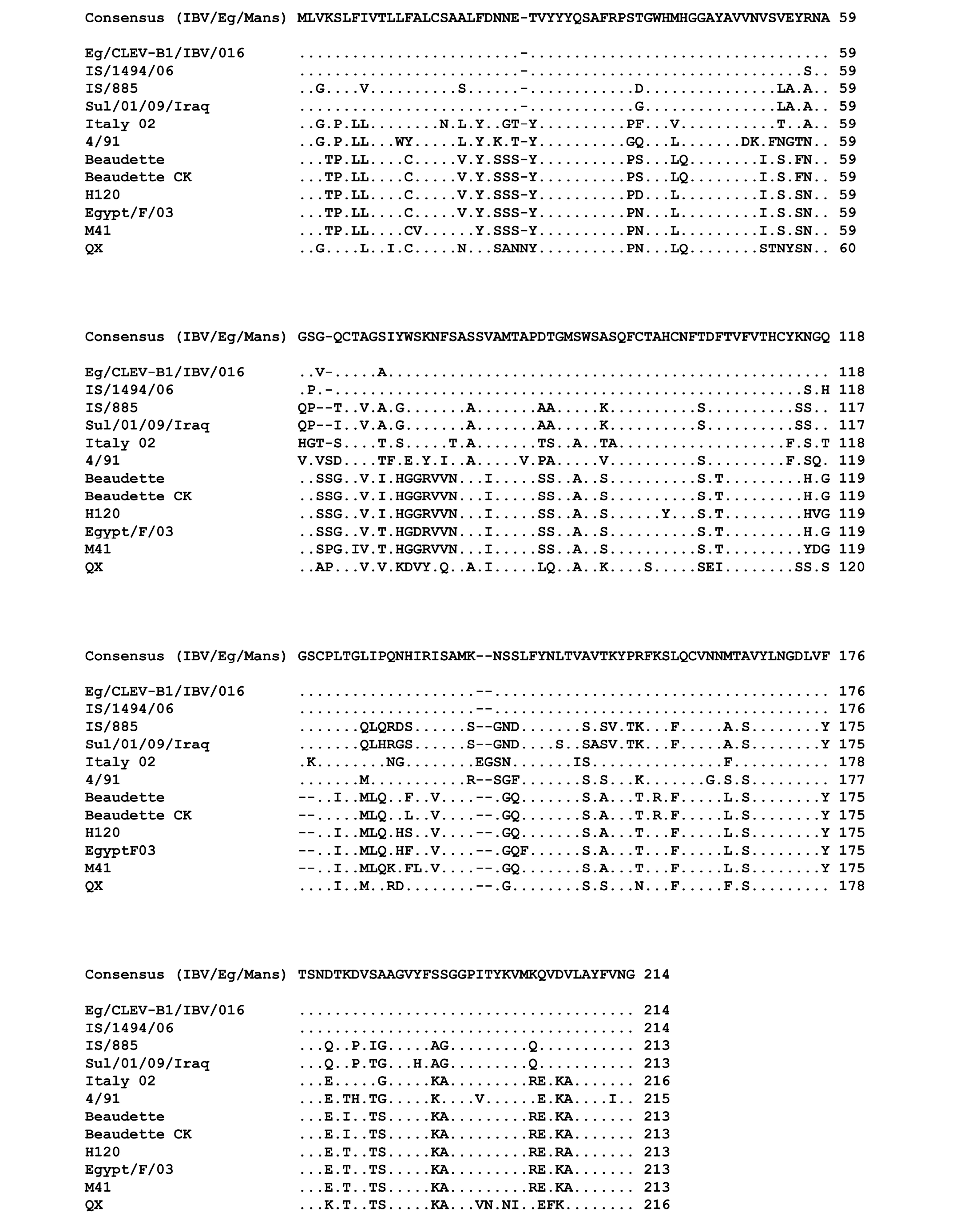
Multiple sequences alignment of the amino acid residues of the partial S-protein sequences for the IBV isolates and selected reference strains
Points indicate amino acid residues that are identical to the consensus sequence, while different amino acid residues are indicated by letters. Gaps which are a result of the alignment are indicated by a minus.
Phylogenetic analysis of the IBV isolates and the reference strains
The partial S-protein sequences on nucleotide (A) and amino acid level (B) of the Egyptian IBV isolates. Scale bars indicate the number of nucleotide /amino acid substitutions per site.
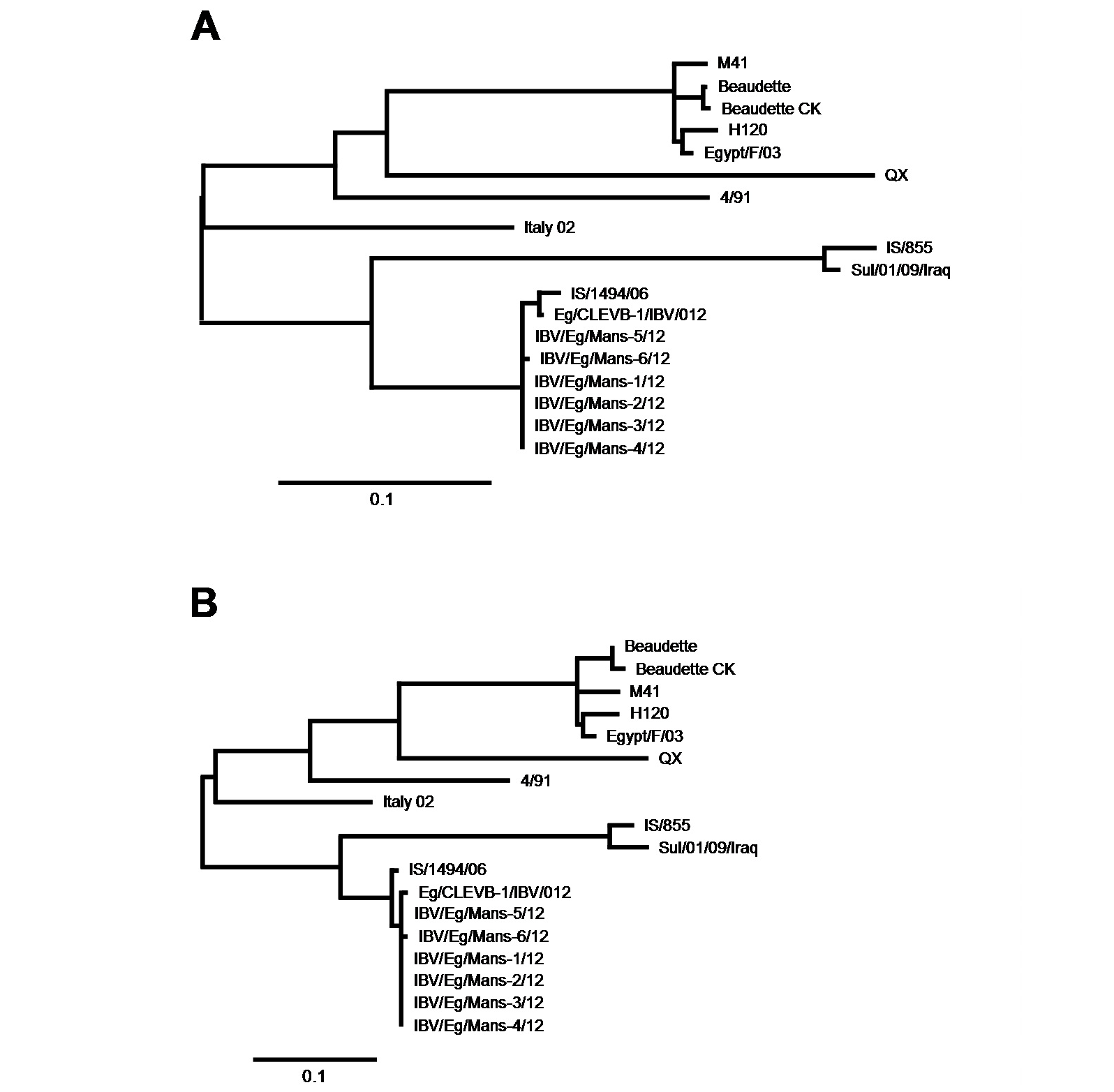
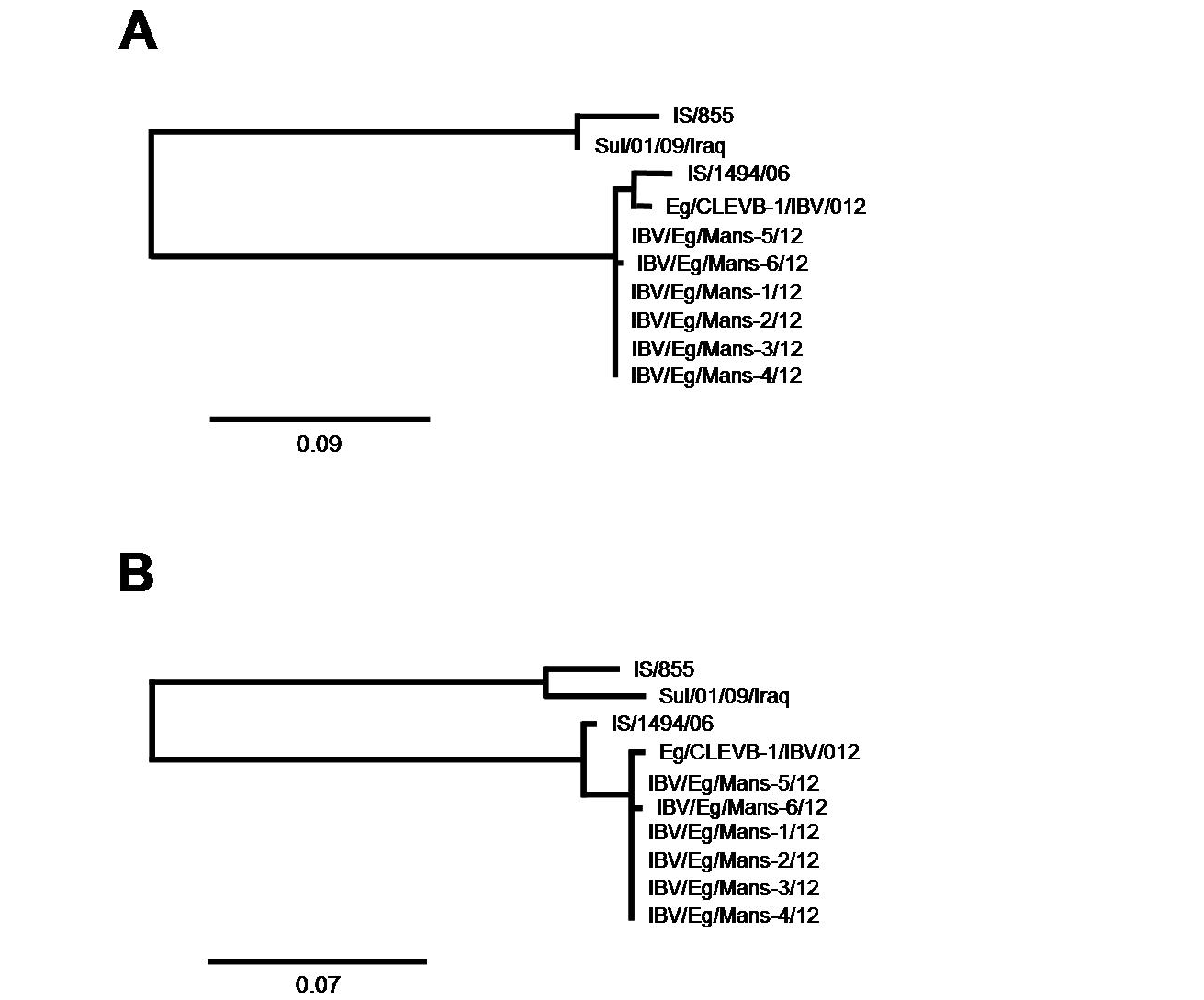
Phylogenetic analysis focusing on closest relatives of the IBV isolates
The partial S-protein sequences on nucleotide (A) and amino acid level (B) of the IBV isolates. Scale bars indicate the number of nucleotide /amino acid substitutions per site.