Advances in Animal and Veterinary Sciences
Research Article
Bacterial Etiology of Skin and Wound Infections along with Antibiogram Profiling in Reference to Emerging Antibiotic Resistance in Animals
Ruchi Tiwari1*, Sharad Kumar Yadav2, Shanker Singh3, Neeraj Kumar Gangwar4
1,2Department of Veterinary Microbiology, 3Department of Veterinary Medicine, 4Department of Veterinary Pathology, Uttar Pradesh Pandit Deen Dayal Upadhayay Pashu Chikitsa Vigyan Vishvidhyalaya Ewam Go-Anusandhan Sansthan (DUVASU), Mathura (U.P.)–281001, India.
Abstract | The present study demonstrated bacterial etiology of skin affections and their antibiogram profiling of different animal species of various age groups. A total of 255 samples from cattle, buffalo, dogs, goats, sheep, camel and horses were collected over a period of two years (2012-2014) from Mathura, (U.P.) India, and nearby surrounding areas. Clinical samples were collected from variety of skin disorders, subjected to laboratory isolation and identification as per standard protocols. The bacterial infection, Staphylococcus aureus, was the most common isolate (36.22 %), followed by E. coli (34.59 %), Pseudomonas (20.54), Bacillus (16.21), Klebsiella (12.43), Micrococcus spp (8.11 %), Streptococcus pyogens (7.56), Proteus (6.49), Clostridium (3.78), Gram negative non-lactose fermenter (2.7%), Gram positive non-spore producing bacilli (2.16) and Fusobacterium (0.54 %). Bacterial isolates obtained were subjected to in vitro antibiotic sensitivity testing by disc diffusion method against 23 commercially available antimicrobial discs. Results of the current study revealed maximum sensitivity to gatifloxacin (94.03%), amikacin (85.7%), mild sensitivity to sparfloxacin (44.85%) and gentamicin (43.48%); emergence of multi-drug resistant bacterial strains of Staphylococcus aureus, Pseudomonas aeruginosa, E.coli appeared to be an important finding. Present study is intended to document the complex microbial inhabitants of wounds from animals and their antimicrobial sensitivity pattern and strongly recommended the use of antimicrobial sensitivity testing to know the rapidly changing pattern of antibiotic activity. The study will further aid to the existing literature for planning new alternative therapeutic strategies against rising global health havoc of multidrug resistance.
Keywords | Bacteria, Skin affections in animals, Wounds, Abscesses, Antibiotic resistance
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | January 12, 2015; Revised | March 27, 2015; Accepted | March 28, 2015; Published | March 31, 2015
*Correspondence | Ruchi Tiwari, College of Veterinary Sciences and Animal Husbandry, DUVASU, Mathura, India; Email: ruchi.vet@gmail.com
Citation | Tiwari R, Yadav SK, Singh S, Gangwar NK (2015). Bacterial etiology of skin and wound infections along with antibiogram profiling in reference to emerging antibiotic resistance in animals. Adv. Anim. Vet. Sci. 3(5): 259-268.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.5.259.268
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Tiwari et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Skin affections including wound infections are frequent complications following lacerations, injuries, penetrating trauma, animal fights, bites etc. In conventional animal rearing practices, wounds were mostly left ignored to be self-cured until they affect the general condition and productivity of the animals including meat, leather quality, or economy of the owner. If treated without confirmatory diagnosis, untargeted therapy and injudicious use of allopathic drugs, gives way to emergence of antimicrobial drug resistant pathogens (Douglas, 1975; Okeke et al., 2005; Tiwari et al., 2013).
Superficial skin infections, which initially appear as painful, red ulcerations are followed by clear to cloudy discharge or may progress as patches of hair loss, redness, and scale (dandruff). Skin ailments or wounds are probably the most common causes of enhanced susceptibility of animals to infections, as they are prone to bacterial contamination. Although prognosis of skin infection is not grave but may adversely affect the working efficiency, quality of hide and occasionally may spread over the body surfaces and may eventually result into internal abscesses, fistula and sometimes septicemia (Tyler et al., 1999).
In veterinary practice, skin affections and inflammation or dermatitis is a common condition especially in small animals and pets and most common causes are bacterial and fungal infections apart from viral and mechanical injuries (Talan et al., 1996). Superficial skin infections are primarily caused by aerobic micro-organism such as Staphylococcus, Streptococcus (Flesh eating bacteria), Vibrio or Aeromonas species, Corynebacteria spp., Pseudomonas aeruginosa, Pasteurella multocida and deeper wounds may be infected with anaerobic pathogens such as Bacteroides, Fusobacterium and Clostridium species. Bacterial infections are typically caused by colonizing normal flora and may also be by antibiotic resistant bacteria such as Methicillin Resistant Staphylococcus aureus (MRSA), Vancomycin Resistant Enterococcus faecium (VREF) and Extended Spectrum Beta lactamases (Watanabe, 1971; Duse, 1998; Boucher and Corey, 2008; Tiwari and Dhama, 2014). Fungal infections may arise later on as they are not inhibited by antibacterial treatment. These conditions are further aggravated by the exposure of nosocomial pathogens and environmental contaminants (Waldron and Zimmerman-Pope, 2003; Wright, 2010). Moreover, traditional animal husbandry practices and conventional rearing facilities with low literacy rate and unawareness of hygienic measures in animal owners further create bottlenecks.
For therapy, antibiotics when used inappropriately and irrationally and course of antibiotic therapy is terminated without prescribed format, favourable conditions for development of resistant group of microbes arise worldwide. In the developing countries nowadays multiple antibiotic resistance have been observed among important pathogens viz., Escherichia coli; MRSA, VREF; Klebsiella; Vibrio etc (Bozdogan et al., 2003; Byarugaba, 2004; Soulsby, 2005; Zhang et al., 2006). Uncontrolled and widespread application of antibiotics in both human and veterinary medicines is equally responsible for increase in prevalence of antibiotic-resistant bacterial infections, and emergence of new antimicrobial resistant pathogenic strains of various infectious agents. Due to paucity of significant work done over animals in this regard, the current study was performed to explore the blueprints of bacterial agents involved in various types of wounds and skin infections (chronic, deep suppurative, open, gun-shot, lacerated, incised, ulcerated wounds etc.) with the history of being unresponsive to earlier treatment in different animal species (cattle, buffalo, goat, horse, camel, sheep, dog) of various age groups, and their sensitivity pattern was assessed against commonly used antibiotics.
MATERIALS AND METHODS
Experimental Population and Sample Collection
A total of 255 samples of wound and skin affections with the history of being unresponsive to earlier treatment were collected from various animal species including cattle, buffalo, goats, sheep, horses, dogs and camel, irrespective of gender and age to screen the cases of superficial wounds and skin infections of animals for bacterial causes from clinical cases and field in and around the Mathura city (Table 1). History of prior antimicrobial or therapeutic treatment was also undertaken. Samples included swabs from wounds of various types viz. horn abscess, gangrenous wound, chronic abscess, deep suppurative wounds, open, gun-shot, lacerated, abrasive, incised, ulcerated, post-operative wounds and wounds over abnormal skin outgrowth. Few samples from skin outgrowth were collected to establish any correlation of abnormal tumorous growth with microbial involvement and subjected to histopathology. All wounds were evaluated independently and accorded infected wound grade. As contaminated wounds may also yield bacterial growth upon culture, hence grading of wounds was made based upon severity of lesion. Lacerated wounds were with irregular edges of more than 10 mm while puncture wounds were wounds less than 10 mm in length approximately. Samples were carefully collected after cleaning the periphery of wound with 70% ethanol swabs. Samples from deep wounds were collected using sterile cotton-tipped swabs (PW003, HiMedia) by taking all aseptic precautions to check the contamination while sample collection. The swab samples were directly submitted to the Department of Veterinary Microbiology, College of Veterinary Sciences, DUVASU University, Mathura (U.P.), India for further laboratory processing by standard procedures.
Table 1: Species-wise distribution of wound sample number
|
Sr No. |
Species affected |
Distribution of wound samples species-wise |
Total number of samples collected |
||
|
Positive |
Negative |
||||
|
Bacteria |
Other agents (Fungi) |
||||
|
1 |
Buffalo |
74 |
1 |
2 |
77 |
|
2 |
Cattle |
29 |
37 |
1 |
67 |
|
3 |
Canine |
52 |
22 |
3 |
77 |
|
4 |
Equine |
14 |
- |
2 |
16 |
|
5 |
Sheep |
6 |
- |
- |
6 |
|
6 |
Goat |
8 |
1 |
1 |
10 |
|
7 |
Camel |
2 |
- |
- |
2 |
|
Total No. of samples |
185/246 (75.80%) |
61/246 (24.80%) |
9 (3.53%) |
255 |
|
|
Percent-wise proportion |
96.47% (246/255) |
3.53% (9/255) |
|||
Isolation and Identification of Bacterial Agents
Collected swab samples were processed for isolation and identification of different aerobic and anaerobic bacterial agents based on the initial screening and characteristics according to the standard operating procedures of the laboratory. Upon receipt as per the history of sample each swab was inoculated into nutrient broth (NB) or Robertson cooked meat (RCM) media and accordingly incubated at 37oC up to 24-48 hrs. under aerobic or anaerobic (in anaerobic jar by excluding the oxygen) conditions, respectively before attempting pure colony isolation over solid nutrient medium. In case of growth, a true representative of each different colony type was sub-cultured and each isolate was identified using phenotypic cultural and morphological identification methods for isolation of pathogenic bacteria. Biochemical tests namely catalase test, oxidase test, carbohydrate fermentation tests, nitrate reduction test, TSI agar slant reaction, MR-VP, indole reaction and Citrate utilization test were carried out for identification of suspected bacteria. Samples are rendered negative if no microbial growth appeared up to 96 hrs. post-inoculation. The generic identification of bacterial isolates was carried out as per the techniques of Cowan and Steel (1975) and further speciation was performed as described by Hungate (1969), Buchanan and Gibbons (1974), Balows et al. (1992), Quinn et al. (1994) and Henry (2001).
Histopathological Study
Samples of abnormal skin outgrowth with lesions of wound were collected for microbiological investigations and in 10% formalin for histopathology. Paraffin–embedded tissues were sectioned at 5μm, processed and stained with Haematoxylin and Eosin. Stained sections were examined under light microscope.
Susceptibility of Bacterial Isolates to Antimicrobial Agents
Recovered bacterial isolates were subjected to antibiotic sensitivity testing against 23 different antimicrobial discs to assess the pattern of antibiotic activity against bacteria in and around Mathura region. The 18 hrs. incubated nutrient broth culture (turbidity equivalent to 3X108 CFU/ml) of isolated bacterial strain was poured on Muller Hinton agar and spreaded uniformly (as per the Clinical and laboratory standards Institute {CLSI} guidelines). Antibacterial discs were applied aseptically to the surface of the plate at an appropriate arrangement with the help of sterile forceps and incubated at 37ºC for 24 hours, aerobically. All the bacteria were subjected to antibiotic sensitivity testing (ABST) against an array of antimicrobial discs ie; Ampicillin (AMP), Amoxycillin, Ampicillin/Sulbactum (A/S), Amikacin (AK), Erythromycin (E), Gentamicin (Gen), Kanamycin (K), Methicillin (MET), Norfloxacin (Nx), Penicillin-G (P-G), Ciprofloxacin (CIP), Chloramphenicol (C), Streptomycin (S), Tetracycline (T), Enrofloxacin (En), Vancomycin (Va), Ceftriaxone (CTR), Cefotaxime (CTX), Cotrimoxazole (Co-T), Gatifloxacine (GAT), Sparfloxacine (SPX), Clindamycin (CD), Colistin (CL) by disc diffusion technique of Kirby-Bauer and results were interpreted as per the standard charts provided from the drug supplier (HiMedia, India) (Bauer et al., 1966; Kahn and Line, 2005; Hindler and Stelling, 2007; CLSI, 2013).
Table 2: Percent-wise distribution of bacterial genera
|
Sr No. |
Name of isolated bacteria |
Total No. of samples examined |
Total No. of bacterial isolates |
Frequency of distribution in % |
|
1 |
Staphylococcus aureus |
185 |
67 |
36.22 |
|
2 |
Streptococcus pyogens |
185 |
15 |
8.11 |
|
3 |
E. coli |
185 |
64 |
34.59 |
|
4 |
Klebsiella |
185 |
23 |
12.43 |
|
5 |
Pseudomonas |
185 |
39 |
21.08 |
|
6 |
Bacillus |
185 |
30 |
16.21 |
|
7 |
Micrococcus spp. |
185 |
15 |
8.11 |
|
8 |
Proteus |
185 |
12 |
6.49 |
|
9 |
Clostridium |
185 |
7 |
3.78 |
|
10 |
Fusobacterium |
185 |
1 |
0.54 |
|
11 |
Gram negative non-lactose fermenter |
185 |
5 |
2.70 |
|
12 |
Gram positive non-spore producing bacilli |
185 |
4 |
2.16 |
RESULT
Wounds break the continuity of the skin and allow organisms to gain access to tissues and cause infection, particularly those that inoculate pathogens into the dermis, frequently cause skin infections. The main cause of open wounds is physical trauma or injury while fighting or bite by another animal, during transportation accidents or due to saddle misfit injuries (Figure 4 and 5). In present study out of 255, 9 samples were rendered negative as no microbial growth appeared till 76 hrs., while 246 samples revealed presence of different species of bacteria and in few cases other agents as fungi (Table 1 and 2). As present study mainly focused on bacterial etiology, other (fungal) agents are not being discussed here, out of 255, 9(3.53%) samples of wounds were negative and 246(96.47%) were positive. Microbiological investigation based upon cultural, morphological and biochemical studies revealed isolation of strains of Staphylococcus aureus, Pseudomonas aeruginosa,
E.coli, Klebsiella spp., Protius, Bacillus, Clostridium, Streptococcus, Micrococcus etc. (Figure 6). The majority of wounds contained a mixture of aerobes and anaerobes, which reflect the diversity of microorganisms present in the atmosphere infecting wounds and vicinity of the affected animal and to a lesser extent of their skin also.
Table 3: Result of antibiotic sensitivity testing of various isolates recovered from wounds of animals
|
Sr No. |
Name of micro organism |
Tot- al No. of isol ates |
Antibiotic discs used (No. of Sensitive isolates) |
||||||||||||||||||||||
|
A M P |
A M C |
A / S |
A K |
E |
G E N |
K |
M E T |
N X |
P - G |
C I P |
C |
S |
T |
E n |
V A |
C T R |
C T X |
Co - T |
G A T |
S P X |
C D |
C L |
|||
|
1 |
Staphyl ococcus aureus |
67 |
1 |
2 |
6 |
22 |
8 |
13 |
0 |
3 |
10 |
0 |
1 |
25 |
15 |
12 |
4 |
6 |
7 |
0 |
0 |
63 |
11 |
0 |
0 |
|
2 |
Strepto coccus pyogens |
15 |
1 |
0 |
0 |
5 |
2 |
3 |
0 |
0 |
1 |
0 |
0 |
4 |
1 |
4 |
0 |
2 |
3 |
0 |
0 |
6 |
3 |
0 |
0 |
|
3 |
E. coli |
64 |
0 |
0 |
8 |
40 |
5 |
25 |
1 |
0 |
4 |
0 |
0 |
26 |
11 |
6 |
3 |
1 |
19 |
2 |
0 |
7 |
11 |
0 |
0 |
|
4 |
Klebs iella |
23 |
1 |
1 |
4 |
8 |
5 |
10 |
0 |
0 |
1 |
0 |
0 |
8 |
1 |
5 |
0 |
1 |
5 |
0 |
0 |
9 |
5 |
0 |
0 |
|
5 |
Pseudo- monas |
39 |
0 |
0 |
1 |
32 |
1 |
15 |
0 |
0 |
6 |
0 |
0 |
21 |
8 |
3 |
2 |
0 |
10 |
0 |
0 |
19 |
9 |
0 |
0 |
|
6 |
Bacill- us |
30 |
0 |
0 |
2 |
17 |
0 |
5 |
0 |
0 |
2 |
1 |
0 |
10 |
3 |
4 |
0 |
1 |
3 |
0 |
0 |
13 |
5 |
0 |
0 |
|
7 |
Micro- coccus |
15 |
0 |
0 |
1 |
7 |
3 |
1 |
0 |
0 |
2 |
0 |
0 |
4 |
2 |
5 |
1 |
0 |
0 |
0 |
0 |
10 |
4 |
0 |
0 |
|
8 |
Prote- us |
12 |
2 |
0 |
3 |
4 |
2 |
4 |
1 |
0 |
0 |
0 |
1 |
4 |
1 |
1 |
0 |
0 |
1 |
0 |
0 |
5 |
3 |
0 |
0 |
|
9 |
Clostri- dium |
7 |
0 |
0 |
0 |
6 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
4 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
|
10 |
Fusob- acteri- um |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
11 |
Gram negative non- lactose fermenter |
5 |
0 |
0 |
0 |
4 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
|
12 |
Gram positive non-spore producing bacilli |
4 |
0 |
0 |
0 |
2 |
0 |
2 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
AMP= Ampicillin; AMC= Amoxycillin; A/S= Ampicillin/Sulbactum; AK= Amikacin; E= Erythromycin; CTX= Cefotaxime; GEN= Gentamicin; CIP= Ciprofloxacin; K= Kanamycin; MET= Methicillin; T = Tetracycline; Nx= Norfloxacin; P-G= Penicillin-G; C= Chloramphenicol; S= Streptomycin; En= Enrofloxaacin; Va= Vancomycin; CTR= Ceftriaxone; Co-T= Cotrimoxazole; GAT= Gatifloxacin; SPX= Sparfloxacin; CD= Clindamycin; CL= Colistin
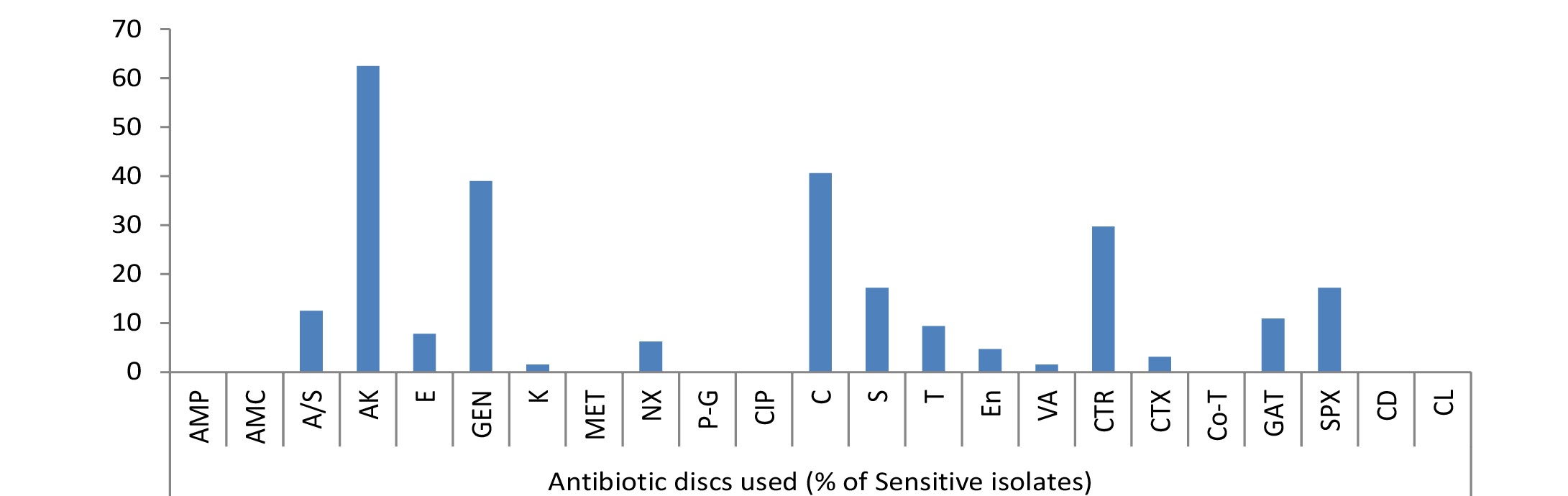
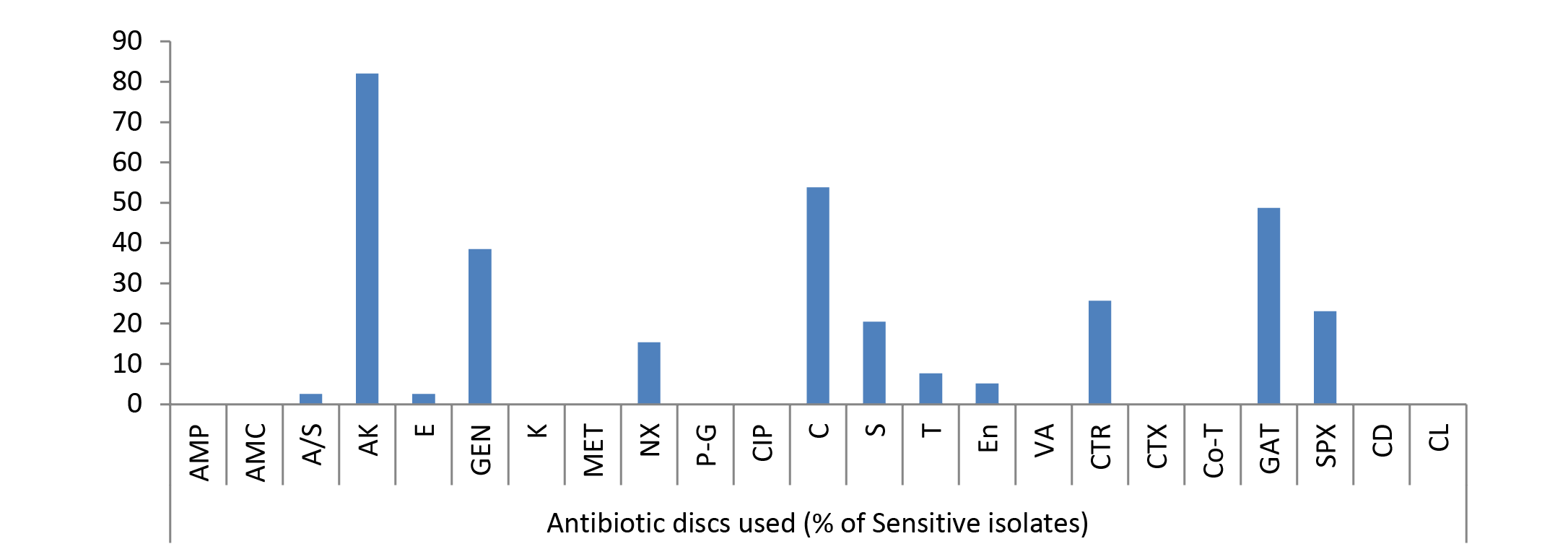
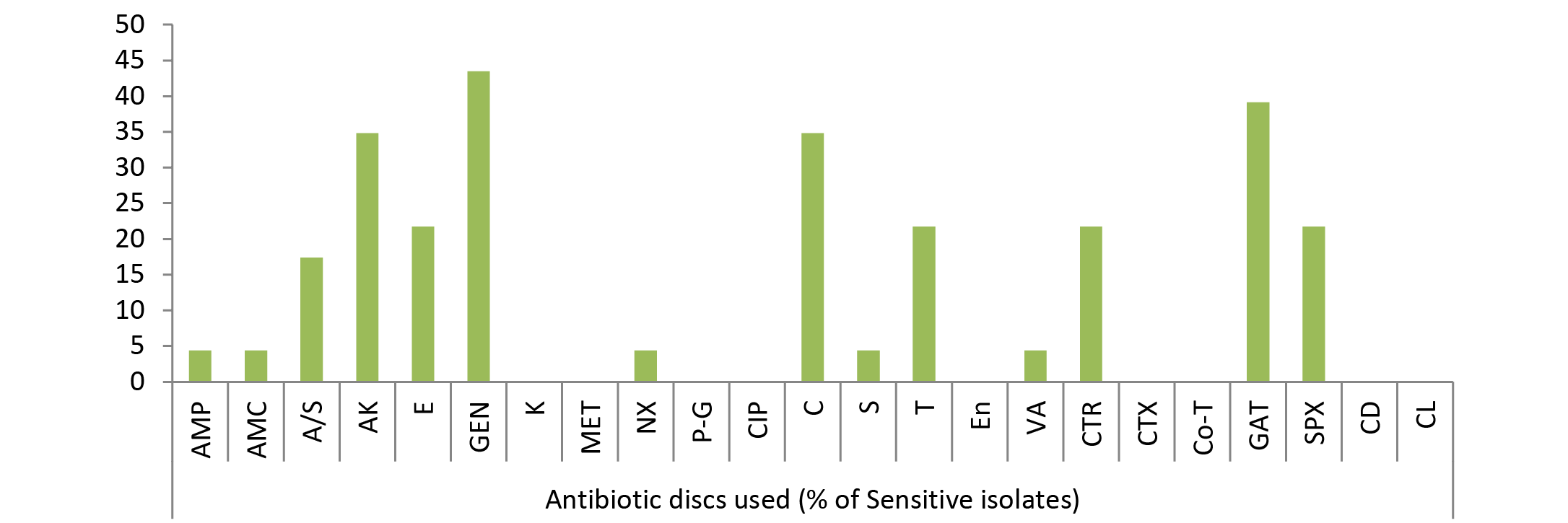
The results of frequency distribution of bacterial isolates were presented in (Table 2). In the study performed over animals for wounds among significant isolates Staphylococcus aureus was the most common isolate (36.22 %), followed by E. coli (34.59 %), Pseudomonas (20.54), Bacillus (16.21) and Klebsiella (12.43). Other bacterial agents involved were less than 10 % as of Micrococcus spp (8.11 %), Streptococcus pyogens (7.56), Proteus (6.49), Clostridium (3.78), Gram negative non-lactose fermenter (2.7%), Gram positive non-spore producing bacilli (2.16) and Fusobacterium (0.54 %). Clostridium spp. and Fusobacterium were the only obligate anaerobes isolated from the samples collected. Bacterial skin infections common in animal veterinary practice, are most frequently caused by Staphylococcus aureus apart from other common causative agents like Escherichia coli as evident from the results. Gram negative bacteria are generally secondary invaders which establishes under chronic conditions. Pseudomonas, however, is a Gram negative notorious bacterium while involvement of Bacillus in 16.21% cases indicated that it is actively causing wound infection rather than present as contaminant because samples were collected under aseptic conditions. Single isolate of Fusobacterium was also identified; one probable reason behind this low prevalence of this microorganism could be good managemental condition on the farm for animal rearing or slow growing tendency of Fusobacterium.
Due to the scarcity of literature on animal studies in this regard present experiment was designed to look in to the pattern of bacterial susceptibility of wound isolates against various antimicrobial drugs. The in vitro antibiotic sensitivity test of different types of all bacterial isolates to 23 different antibiotics such as Ampicillin, Amoxycillin, Ampicillin/Sulbactum, Amikacin, Erythromycin, Gentamicin, Kanamycin, Methicillin, Norfloxacin, Penicillin-G, Ciprofloxacin, Chloramphenicol, Streptomycin, Tetracycline, Enrofloxacin, Vancomycin, Ceftriaxone, Cefotaxime, Cotrimoxazole, Gatifloxacine, Sparfloxacine, Clindamycin and Colistin was studied. A slight variation was noticed in the results of sensitivity of isolates against 23 different antibiotics used, though no single antibiotic therapy was found effective against all the bacteria. Antibiotic sensitivity pattern of most of bacteria, isolated from different cases of skin/wound infections, revealed resistant strains even against multiple drugs. Gatifloxacin, sparfloxacin, amikacin, chlloramphenicol and gentamicin were highly sensitive drugs in most of the cases (Figure 1, 2 and 3). The isolated bacteria were less sensitive to erythromycin, colistin, cefotaxime, Enrofloxacin, Vancomycin, Tetracycline, Ceftriaxone, Clindamycin, Streptomycin, and Kanamycin while resistant to Ampicillin, Amoxycillin, Ampicillin/Sulbactum, Erythromycin, Methicillin and Norfloxacin in most of cases (Table 3).
Table 4: Sample of wounds over abnormal skin/tissue outgrowth
|
Sr No. |
Species examined for abnormal skin outgrowth with wound over it |
No. of swab samples for microbiological investigation |
Total No. of sample |
|
|
Positive |
Negative |
|||
|
1 |
Cattle |
1 |
1 |
2 |
|
2 |
Buffalo |
3 |
1 |
4 |
|
3 |
Canine |
2 |
3 |
5 |
|
4 |
Equine |
--- |
1 |
1 |
Figure 4: Gun-shot wound in buffalo
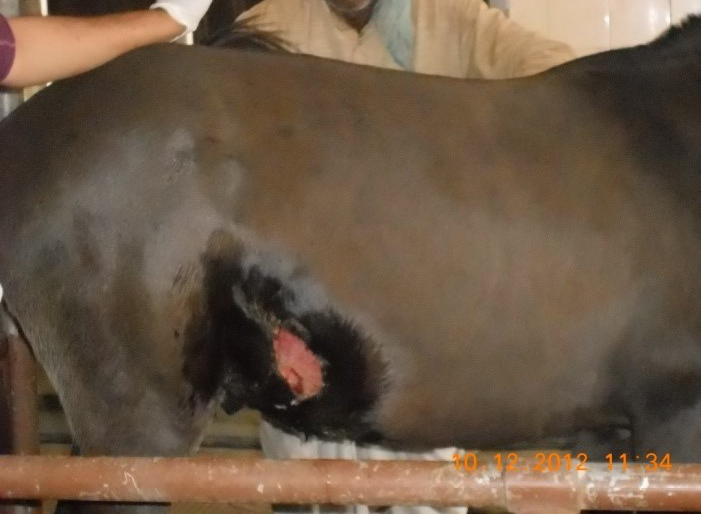
Figure 5: Lacerated wound in horse
Among Streptococcus pyogens 40% isolates against gatifloxacin and 33.3% against amikacin were sensitive, rest 26.67% were mild sensitive to chloramphenicol and tetracycline, 13.33% to erythromycin and vancomycin, 6.67% to ampicillin, norfloxacin and streptomycin, 4.48% to gentamicin, ceftriaxone, sparfloxacin while no strains were sensitive to amoxycillin, ampicillin/sulbactum, kanamycin, methicillin, penicillin-G, ciprofloxacin, enrofloxacin, cefotaxime, cotrimoxazole, clindamycin and colistin.
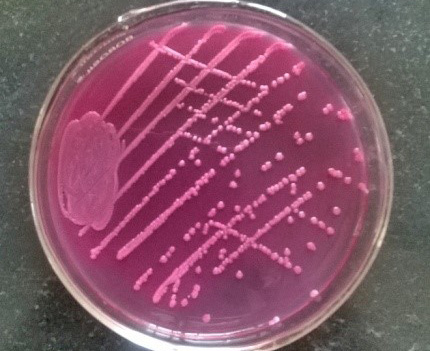
Figure 6: Pink colonies over MLA
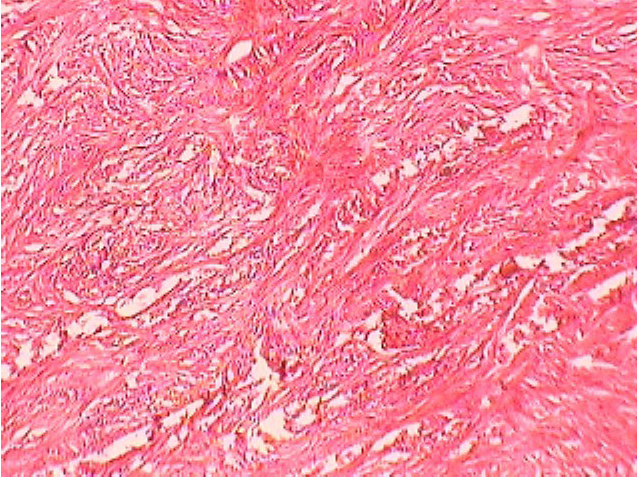
Figure 7: Fibro-sarcoma in Buffalo
Out of 34.59% E. coli isolates, maximum sensitivity was shown by 62.5% strains against amikacin but no other drug showed sensitivity of more than 50%. Among rest isolates 40.62% to chloramphenicol, 39% to gentamicin, 29.69% to ceftriaxone, 17.19% to streptomycin and sparfloxacin, 12.5% to ampicillin/sulbactum, 10.94% to gatifloxacin, 9.38% to tetracycline, 7.81% to erythromycin, 6.25% to norfloxacin, 4.69% to enrofloxacin, 3.12% to cefotaxime, 1.56% to kanamycin and vancomycin were mild sensitive while ampicillin, amoxycillin, methicillin, penicillin-G, ciprofloxacin, cotrimoxazole, clindamycin and colistin were resistant drugs (Figure 1).
Out of 23 isolates of Klebsiella spp., 43.48% were sensitive to gentamicin; 39.13% to gatifloxacin; 34.78% to amikacin, chloramphenicol; 21.74% to sparfloxacin, ceftriaxone, tetracycline, erythromycin; 17.39% to ampicillin/sulbactum, 4.35% to ampicillin, amoxycillin, norfloxacin, streptomycin, vancomycin (Figure 3) while kanamycin, methicillin, penicillin-G, ciprofloxacin, enrofloxacin, cefotaxime, cotrimoxazole, clindamycin and colistin drugs failed to show sensitivity as no zone of inhibition was produced against these antimicrobial discs.
Amongst 39% Pseudomonas spp. isolates recovered from wound cases 82.05% strains showed maximum sensitivity to amikacin followed by 53.84% strains sensitive to chloramphenicol (Figure 2). Rest of drugs showed mild sensitivity, 48.72% isolates sensitive to gatifloxacin; 38.46% to gentamicin; 25.64% to ceftriaxone; 23.07% to sparfloxacin; 20.51% to streptomycin; 15.38% to norfloxacin; 7.69% to tetracycline; 5.13% to enrofloxacin; 2.56% to erythromycin, ampicillin/Sulbactum; and ampicillin, amoxycillin, kanamycin, methicillin, penicillin-G, ciprofloxacin, vancomycin, cefotaxime, cotrimoxazole, clindamycin and colistin were resistant.
As evident from Table 3 result of current study illustrated that among isolated bacteria Staphylococcus aureus, Streptococcus pyogens, Micrococcus and Proteus were found most sensitive against gatifloxacin; E. coli, Bacillus, Pseudomonas, Clostridium against amikacin and Klebsiella against gentamicin. Single isolate of Fusobacterium was sensitive against streptomycin, Gram negative non-lactose fermenters showed maximum sensitivity to amikacin while Gram positive non-spore producing bacilli were most sensitive to amikacin, gentamicin and tetracycline.
Histopathology and microbiological investigation suggested bacteria were not the route cause in cases of abnormal skin growth with wounds (Figure 7). Bacterial agents were involved as secondary agents in 50% cases (Table 4). Out of 12, 6 (50%) samples were negative for presence of any microbial agent while from other 6 cases (50%) Staphylococcus epidermidis, Bacillus spp. Gram negative non-lactose fermenter and Gram positive non-spore producing bacilli could be isolated. Certain carcinogens or continuous irritant could be the possible reason behind development of abnormal tissue growth over a long period of time. Involvement of viral agent was beyond the scope of present study, moreover no spread in other parts of body further ruled out the possibility of any infectious agent as etiology. Histopathology suggested fibrosarcoma, fibroma, squamus cell carcinoma and venereal sarcoma from different abnormal tissue outgrowth which could be due to any consistent irritant or action of any carcinogen.
DISCUSSION
Present study mainly focused on screening of bacterial agents involved in cases of skin infections, wounds and from abnormal skin growth from variety of animal species which were unresponsive to earlier treatments. Microbiological investigation revealed isolation of strains of Staphylococcus aureus, Pseudomonas aeruginosa, E.coli, Klebsiella spp., Protius, Bacillus, Clostridium, Streptococcus, Micrococcus and Fusobacterium from different wounds from various species. Our result of bacterial isolation is in support with earlier reports of other workers (Goldstein, 1992; Kelly et al., 1992; Jousmies-Somer et al., 1995; Qureshi et al., 2002; Dierikx et al., 2012; Escobar et al., 2013). In the present study performed over animals for wounds among significant isolates Staphylococcus aureus was the most common isolate (36.22%) followed by E. coli from 34.59 % cases and showed different pattern of sensitivity as evident from the Table 2 and 3. Kikuvi et al. (2007) also reported resistance against streptomycin and chloramphenicol in Escherichia coli recovered from cattle and poultry in Kenya. In our results also only 17.19% isolates were sensitive to streptomycin. Findings of Albrechtova et al. (2012) described that dogs may act as reservoir of resistant E. coli strains against cephalosporins and further may spread the resistant genes in the population. They are in accordance to our results of ABST pattern of E. coli as most of strains were refractive to antibiotics of cephalosporin group. Similarly, Rubin and Pitout (2014) have reported resistant strains of E. coli from cat, dogs and horses in their study.
Chopra and Roberts (2001) and Hawkey and Jones (2009) documented in separate studies that antibiotic sensitivity testing of the bacterial agents, isolated from different cases of skin/wound infections, revealed resistant strains even against multiple drugs. In the VET 01-A4 section of CLSI document (2013) instructions regarding routine antimicrobial sensitivity testing, proper reporting and recommendations about antimicrobial agents to be used specifically in domestic and companion animals is described in detail in order to have a watch on emerging antimicrobial resistance (AMR) against various bacterial agents. Alike to the reports of Chopra and Roberts (2001) and Hawkey and Jones (2009) in our recent study also resistance against multiple drugs was noticed.
In current study, among isolated bacteria Streptococcus pyogens, Staphylococcus aureus, Micrococcus and Proteus were found most sensitive against gatifloxacin; E. coli, Bacillus, Pseudomonas, Clostridium against amikacin, Klebsiella against gentamicin and Fusobacterium was found most sensitive against streptomycin, however they showed resistance against multiple other drugs as evident from the Table 3 and Figure 1, 2 and 3. The extensive antimicrobial susceptibility to Gatifloxacin in the current study may be due to their infrequent use in veterinary practice. The use of most fluoroquinolones (ciprofloxacin) and the aminoglycosides (streptomycin) in majority of cases even without confirmatory diagnosis of cause could be one possible reason behind rapidly developing resistance against the drugs of these classes (Kopecko and Punch, 1971; Williamson, 1983; von Baum and Marre, 2005; Ewers et al., 2012; Escobar et al., 2013). Irrational and indiscriminate use of antibiotic drugs led to unresponsiveness of microbial agents or developing resistance of bacterial strains against various drugs. The development of antibiotic resistance among the bacteria is an important issue for treatment of infectious diseases in man as well as animals. Although, there is progress in research and development of new and improved salts of antimicrobial agent but the bacteria are developing resistances to these antibiotics at a higher speed from it. The antibacterial resistance observed in the isolated bacterial strains might be due to arbitrary use of those antibacterial agents in field or incomplete follow-up of treatment in nearby rural as well as urban areas of Mathura. Results are reminiscent of interspecies transmission between humans and companion or domestic animals due to close proximity as one possible reason for spread of resistant bacterial strains. Molecular studies are required to further confirm any chromosomal mutation and the presence of episome or specific plasmid and its horizontal or clonal transfer in the environment (Watanabe et al., 1964; Aminov and Mackie, 2007; Allen et al., 2010).
Histopathology results suggested that no microorganism was the cause of abnormal tissue growth in current study. These results were in contrast with the findings of Antuofermo et al. (2014), who isolated Mycobacterium chelonae from tumor like skin outgrowth and oral masses from fishes, though not from domestic animals. In another study, Guarner and Brandt (2011) described that histopathological techniques are good for the diagnosis of systemic fungal infections. Guarner (2012) reported presence of microorganisms in formalin fixed granulomas by histopathology, his findings were different than the results recorded in the current study but yet literature is sparsely available exploring involvement of bacteria in abnormal skin outgrowth in pet and domestic animals.
CONCLUSIONS
The present paper highlighted the complex microflora involved in various wounds and their emerging antimicrobial resistance pattern against various antibiotic drugs. In veterinary practice, skin affection and wound infection are the most frequent complication. The results of the current study projected that animal wounds entail specific diagnostic fact sheet to support veterinary assistance for speedy and well-in-time recovery. Due to the paucity of animal studies in this regard, present paper significantly described and fills the gap in knowledge that screening of superficial wounds and skin infections of 255 samples from cattle, buffalo, goats, camel, sheep, horses and dogs from Mathura and nearby surrounding areas revealed involvement of bacteria (75.20 %) as causative agents. Microbiological investigation revealed isolation of strains of Staphylococcus aureus, Pseudomonas aeruginosa, E.coli, Klebsiella spp., Proteus, Bacillus, Clostridium, Streptococcus, Micrococcus, Fusobacterium, Gram negative non-lactose fermenter, Gram positive non-spore producing bacilli in different percentage. Antibacterial resistant strains of Staphylococcus aureus, multi-drug resistant strains of Pseudomonas aeruginosa, E.coli, Klebsiella spp were observed. Gatifloxacin, amikacin, chloramphenicol and gentamicin were highly sensitive drugs in most of cases. Bacteria were not route cause in cases of abnormal skin growth. The sensitivity tests performed would definitely provide guideline to the veterinarian to select appropriate antibiotics to reduce the economic losses through selecting the sensitive antibiotics. The findings of the present study concluded that the rapid emergence of resistant bacterial isolates from wounds of animals accentuates the potential impact of antimicrobial resistant organisms and is a call to action for the investigation of molecular basis of drug resistance in companion domestic animals and other veterinary species. Molecular studies are required to further confirm any chromosomal mutation and the presence of specific plasmid or episome and its horizontal or clonal transfer in the environment. Study further warrants the effective implementation of rapid diagnostic for appropriate prevention and control measures against important emerging multidrug resistant bacteria.
ACKNOWLEDGEMENT
The authors cordially thank Dean, College of Veterinary Sciences & Animal Husbandry, DUVASU, Mathura (U.P.) for his scientific advice, proof reading the manuscript and acknowledge the help for administrative financial support and institutional research funding.
CONFLICT OF INTEREST
The authors declare no competing interests.
REFERENCES