Advances in Animal and Veterinary Sciences
Research Article
Phage Immobilized Antibacterial Silica Nanoplatform: Application against Bacterial Infections
Neha Bhardwaj1, 2, Sanjeev Bhardwaj1, 2, Pooja Devi1, Swati Dahiya2*, Madan Lal Singla1, Chirravoori Ghanshyam1, Minakshi Prasad1*
1Agrionics, CSIR-Central Scientific Instruments Organisation, Chandigarh-160030, India; 2University Institute of Engineering and Technology, Kurukshetra University, Kurukshetra-136119, India.
Abstract | Antibiotic resistance in bacterial pathogens has become a worldwide public health problem in veterinary and human medicine. The emergence of methicillin resistance in Staphylococcus sp. has led to complications in treatment of bovine mastitis, wound infections in horses, dogs, pigs etc. and bacterimia in humans ranging from postules, sepsis to even death. To address this problem of antibiotic resistance, several attempts are being made of which, phage therapy offers a promising alternative. Herein, we report a phage based antibacterial nanoplatform having bactericidal potential against an isolated methicillin resistant skin bacterium, Staphylococcus arlettae as a model organism. The bacteriophages (phages) used in the study were isolated from sewage water via enrichment method against an isolated methicillin resitant skin bacterium, Staphylococcus arlettae (GenBank accession no. KF589201.1). The isolated phages were characterized thereafter for their host specificity and morphology by plaque assay and transmission electron microscopy (TEM) respectively. Intrinsic charge characteristic of phages was utilized for their immobilization on silica NPs monolyer using different conjugation chemistry, through covalent/electrostatic/ physisorption interactions. The efficacy of each immobilization approach was compared using plaque assay. Silica NPs of size ~ 300 nm were synthesized by hydrolysis and condensation of tetraethylorthosilicate in an alkaline environment, and characterized using scanning electron microscopy (SEM). The particles were dispersed at air/water interface for monolayer formation on polyvinyllidine fluoride (PVDF) membrane via Langmuir-Blodgett (LB) thin film deposition technique at 15 mN/m surface pressure. It was observed that 3-aminopropyltrimethoxysilane (APTMS) treated membranes act as a better bactericidal platform due to proper orientation of phages by means of electrostatic and covalent interaction as compared to phage bound by physical adsorption. Moreoever, high surface/volume ratio of NPs further helped in improving bactericidal performance, associated with large loading capacity of phages on film. Thus, the present study can facilitate in design and development of an effective anti-bacterial compositions and dressings in veterinary medicine and animal agriculture to combat Staphylococcal infections.
Keywords | Silica nanoparticles, Langmuir Blodgett Film, Phages, Anti-bacterial
Editor | N. K. Mahajan, Head of the Department, Department of Veterinary Public Health and Epidemiology, LUVAS, Hisar, Haryana, India.
Special Issue | 1(2015) “Biotechnological and molecular approaches for diagnosis, prevention and control of infectious diseases of animals”
Neha Bhardwaj, Sanjeev Bhardwaj and Pooja Devi contributed equally for this paper.
Received | December 01, 2014; Revised | March 18, 2015; Accepted | March 21, 2015; Published | March 30, 2015
*Correspondence | Swati Dahiya, Department of Vetrinary Microbiology, Lala lajpat Rai University of Vetrinary and Animal Sciences, Hisar, Haryana, India; Email: swatidahiya@yahoo.com; Minakshi Prasad, Department of Animal Biotechnology, Lala lajpat Rai University of Vetrinary and Animal Sciences, Hisar, Haryana, India; Email: minakshi.abt@gmail.com
Citation | Bhardwaj N, Bhardwaj S, Devi P, Dahiya S, Singla ML, Ghanshyam C, Minakshi P (2015). Phage immobilized antibacterial silica nanoplatform: application against bacterial infections. Adv. Anim. Vet. Sci. 3(1s): 1-9.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.1s.1.9
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Bhardwaj et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Staphylococcus sp. is an important gram-positive bacterial pathogen responsible for several infections in animals such as soft tissue abscesses, wound infections, endocarditis, osteomyelitis, bacteremia, mastitis etc. and thus, is a cause of great concern worldwide. In addition, the emergence of antibiotic resistance in Staphylococcus sp. (particularly methicillin resistance) and ability to persist undetected in herds further apprehends the need for their control measures (DeGraves and Fetrow, 1993; Mann, 2008). Staphylococci are associated with skin diseases in horses such as acne and furunculosis which occur on irritated or damaged skin areas due to friction of a saddle or harness (Smith, 1958). In dairy animals, Staphylococcus sp. is associated with mastitis disease even in countries using most advanced technical dairying practices (McCoy, 1960). Staphylococcal infections are also prevalent in swine conditions such as impetigo, acne, botryomycosis etc. and in pet animals such as dogs where infection follows some form of injury (Hoskins, 1959). It has been reported that animal species, including the food animals and pets are a common source of human MRSA (Methicillin Resistant Staphylococcus aureus) infections, especially in veterinarians who work closely with animals (Weese, 2010).
Bacteriophages have emerged as promising alternative to antibiotics and have shown to be one of the highly efficient and effective solutions for controlling Staphylococcal infections as their life cycle is connected exclusively with bacterial cells (Banowski and Simmering, 2011; Donovan, 2007). These are stable macromolecular assemblies, which can maintain their ability to infect for decades, and are relatively insensitive to wide range of temperature, pH and ionic strength (Pearson et al., 2013; Kutter and Sulkvelidze, 2005). The antibacterial acitivity of bacteriophages has been exploited for their applications as sanitizers, disinfectants, food packagings and in detection of bacterial pathogens (Shin et al., 2011; Singh et al., 2009; Yang et al., 2013). It has been reported by various researchers that the oriented immobilization of bacteriophages (with their tails free for bacterial binding) on a substrate can aid in development of a highly active biosorbent that can simultaneously capture and lyse the target bacterium with high specificity and in close to real-time format (Anany et al. 2011; Singh et al., 2009; Yang et al., 2013). Several studies have been reported regarding immobilization of phages on various substrates such as glass, gold, polytetrafluoroethylene, polyethylene, paper, cellulose, etc. (Anany et al. 2011; Pearson et al., 2013; Singh et al., 2009; Yang et al., 2013; Tawil et al., 2013). Also, the number of phages immobilized on a surface can be increased using nanoparticles as it is a well known-fact that as the size of a particle decreases, the ratio of the surface area to the volume of the particle increases, causing a large number of the entities to reside on the surface (Bashar et al., 2013). Further, the nanoparticles can be deposited on various substrates using different techniques such as drop casting, dip coating, spin coating, etc (Hutter and Maysinger, 2013; Wang et al., 2008). However, Langmuir-Blodgett (LB) based depostion technique offers advantage of precise high level control of (mono and multi) thickness and enables homogenous deposition of monolayer/film over a large area with controlled composition and reproducibility, hence increases the overall surface area available for biomolecule binding.
Thus, in the present work, the bacteriophages have been conjugated to silica nanoparticle’s LB monolayer over a polyvinylidine diflouride (PVDF) membrane as silica nanoparticles offer benefits of easy preparation, good biocompatibily and simple tailorble surface chemistry (He and Hu, 2004). In the study, bacteriophages were first isolated and characterised against one of the methicillin resistant bacterial isolates from human skin, identified as Staphylococcus arlettae and their bactericidal activity was confirmed using plaque assay. Further, the phages were conjugated to silica nanoparticles (NPs) Langmuir-Blodgett (LB) film in an oriented manner using different surface chemistries and their bactericidal effectiveness was then investigated in comparison to random immobilisation by physical adsorption method.
Materials and Methods
All the chemicals used were of highest purity grade. The chemicals used were Soya Casein Digest (SCD) agar, SCD medium, nutrient agar, bacteriological agar powder, Gram staining kit, mannitol salt agar, Phosphate Buffered Saline (PBS, pH = 7.2), Tris buffer (pH= 7), sodium chloride, calcium chloride, glucose, antibiotic sensitivity discs (obtained from HiMedia Laboratories) and ferric chloride, magnesium sulphate (obtained from Central Drug House Laboratories). Membrane filters (pore size = 0.22 μm) used for sterilization of buffers and purification of phages were purchased from Millipore. Tetraethyl orthosilicate (TEOS), ammonium hydroxide solution, ethanol, chloroform, 3-aminopropyl trimethoxy silane (APTMS), (N-(2-aminoethyl)-3-aminopropyl)trimethoxysilane (AEPTMS), cetyltrimethylammonium bromide (CTAB) and gluteraldehyde purchased from Sigma Aldrich were used in the study.
Isolation and Characterization of Bacteria and its Specific Phages
Isolation and Characterisation of Bacteria
The samples for bacterial isolation were collected from the human skin by swabbing PBS dipped sterile cotton, which were then serially diluted and plated onto SCD agar media (Prescott et al., 2002a). After 24 hrs. of incubation, visible bacterial colonies on agar plate were picked and their pure cultures were prepared in the form of slants. Six of the appeared bacterial isolates namely, SDKU, SDKU2, SDKU3, SDKU4, SDKU5 and SDKU6, were characterized by Gram Staining and their ability to grow on selective agar plates was assessed. One of the isolates, SDKU6, was further characterized by ribotyping (Applied Biosystems genetic analyzer 3130, Life Technologies, Germany) using forward primer, RW01, 5’-AAC TGG AGG AAG GTG GGG AT-3’, and reverse primer, DG74, 5’-AGG AGG TGA TCC AAC CGC A-3’, to get genus and species level information (Stackebrandt and Goodfellow, 1991). The sequence so obtained was then analyzed using BLAST in NCBI database and submitted to GenBank (GenBank accession no. KF589201.1) (Altschul et al., 1990). Also, the bacteria SDKU6 and SDKU were sent to Institute of Microbial Technology (IMTECH), Chandigarh, a premier CSIR laboratory of India, for phenotypic characterization and identification.
The characterized and identified bacterial isolates (SDKU and SDKU6) were also tested for their senstivity against several antibiotics (listed in Table 1) by standard disk diffusion method (Koneman et al., 1997). A sterile cotton swab was dipped into the culture suspensions (overnight grown) of bacteria SDKU and SDKU6 confirmed from IMTECH and spread on the surface of sterile SCD agar plates. Standard commercially prepared, fixed concentration antibiotic discs (HiMedia) were placed on the surface of the seeded agar plates. The plates were then incubated for 24hr at 37°C for the formation of zones of inhibition.
Table 1: List of Antibiotics used in the study
|
S. No. |
Antibiotic |
Symbol |
|
1. |
Gentamycin |
G10 |
|
2. |
Vancomycin |
VA30 |
|
3. |
Ofloxacin |
OF5 |
|
4. |
Tetracycline |
T30 |
|
5. |
Amoxycillin |
AM25 |
|
6. |
Methicillin |
M5 |
|
7. |
Clindamycin |
CD2 |
|
8. |
Amphotericin-B |
AP20 |
|
9. |
Chloramphenicol |
C30 |
|
10. |
Erythromycin |
E15 |
Isolation and Characterisation of Bacteriophages
The isolation of bacteriophages against one of the bacterial isolates, SDKU6, was carried out by enrichment method using sewage water samples collected from various sites of Kurukshetra, India (Jordan et al., 2011). In a simple procedure, the water samples were collected and first enriched for bacteriophages against SDKU6. Then, phages were isolated by double agar overlay technique. For this, infection mixture containing 500 µl of enriched sample filtrate and 300 µl of overnight grown host bacterium was prepared, mixed with molten top agar (SCD medium containing 0.6% agar, 5 ml), and quickly poured over sterile SCD agar plates (prepared earlier). The plates were gently swirled and incubated at 37º C for a period of 24 to 48 h to observe clear zones (plaques) on plates. Individual plaques were picked using sterile micropipette tip in PBS buffer and centrifuged at 5000 rpm for 10 min to pellet down the agar particles and bacterial residues. The resulting supernatant containing phages was filter sterilized through 0.22µ Millipore filter and ultracentrifuged (Sorvall WX ULTRA series, Thermo Scientific) at 50,000 rpm for 1h at 4ºC. The titre of purified phages was determined by double agar overlay method (Jordan et al., 2011).
The purified phages were then characterized for their host specificity using plaque assay against various bacterial strains namely, Staphylococcus aureus (MTCC, 3160), Staphylococcus epidermidis (MTCC, 435), Streptococcus mutans (MTCC, 1943), Escherichia coli (MTCC, 443), Pseudomonas aeruginosa (MTCC, 3163), along with the isolated bacterial strain, SDKU6 (Fan et al., 2012). Further, the phages were morphologically characterised by TEM (JEOL, JEM 2100, USA) (Synnott et al., 2009).
Synthesis and LB Film Formation of Silica NPs
Silica NPs were synthesized following an earlier reported method and then characterized for their size, morphology and dispersity using scanning electron microscope (SEM, Hitachi S-4300) (Devi et al., 2013). For LB film formation, an appropriate amount (624 µl) of the silica suspension (10 mg/ml) in chloroform was spread on top of pure water sub-phase using a micro-syringe (1705 N, Hamilton Co.), and a waiting period of 20 min was taken for solvent evaporation. The dispersed particles at the air/water interface were continuously compressed at a barrier speed of 4 mm min−1 and the consequent isotherm was monitored using KSV NIMA LB system. The monolayer formed at air/water interface was transferred onto PVDF membrane and then air-dried before immobilization of bacteriophages.
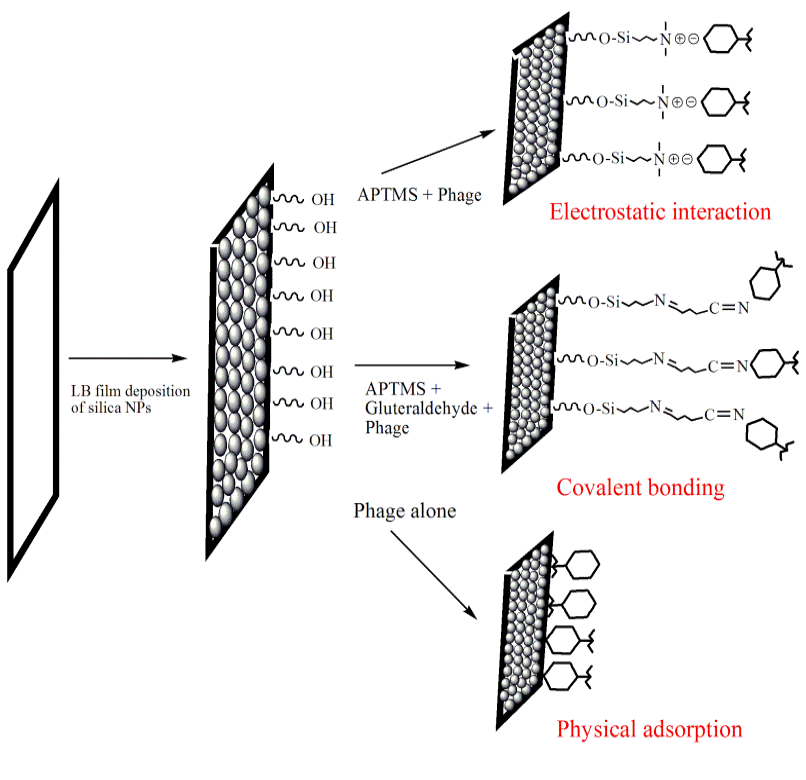
Figure 1: Schematic of effect of binding interactions (electrostatic, covalent and physiosorption) on phage orientation while immobilization on Silica NPs LB film substrate
Table 2: Approaches used for modification of silica NPs film
|
Sample |
Substrate |
Modifying Agent |
Nature of Interaction |
Concentration of Modifying agent (moles/L) |
|
S1 |
PVDF |
Silica LB + APTMS + Phage |
Ionic |
0.011 |
|
S2 |
PVDF |
Silica LB + APTMS + Gluteraldehyde + Phage |
Covalent |
0.011 |
|
S3 |
PVDF |
Silica LB + AEPTMS + Gluteraldehyde + Phage |
Covalent |
0.011 |
|
S4 |
PVDF |
Silica LB + AEAPTMS + Phage |
Ionic |
0.011 |
|
S5 |
PVDF |
Silica NPs LB + Phage |
Physisorption |
- |
|
S6 |
PVDF |
Silica NPs LB + CTAB |
Positive control |
0.011 |
|
S7 |
PVDF |
Phage alone |
Physisorption |
- |
|
S8 |
PVDF |
- |
Negative control |
- |
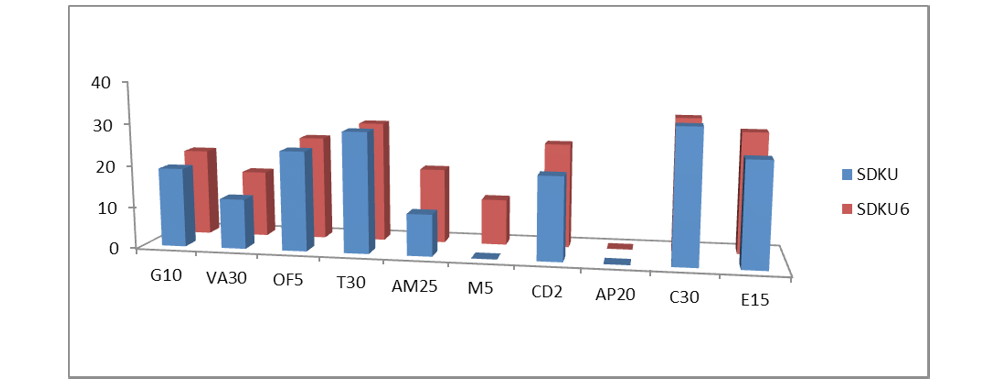
Figure 2: Susceptibility of bacteria Staphylococcus arlettae SDKU and SDKU6 against various antibiotics
Table 3: Characterisation of isolated bacterium SDKU6
|
Antibiotic |
Zones of Inhibition (mm) |
|
|
Staphylococcus arlettae SDKU |
Staphylococcus arlettae SDKU6 |
|
|
Gentamycin G10 |
19 (Susceptible) |
21 (Susceptible) |
|
Vancomycin VA30 |
12 (Resistant) |
16 (Susceptible) |
|
Ofloxacin OF5 |
24 (Susceptible) |
25(Susceptible) |
|
Tetracycline T30 |
29(Susceptible) |
29(Susceptible) |
|
Amoxycillin AM25 |
10 (Resistant) |
18(Intermediate) |
|
Methicillin M5 |
0 (Resistant) |
11 (Resistant) |
|
Clindamycin CD2 |
20 (Susceptible) |
25 (Susceptible) |
|
Amphotericin-B AP20 |
0 (Resistant) |
0 (Resistant) |
|
Chloramphenicol C30 |
32 (Susceptible) |
32 (Susceptible) |
|
Erythromycin E15 |
25 (Susceptible) |
29 (Susceptible) |
Immobilization of Phages onto Silica NPs LB Film
The purified phages were immobilized onto silica NPs Langmuir-Blodgett (LB) film using various conjugation chemistries (schematically shown in Figure 1) after modifying them with different chemicals (S1-S8, Table 2). LB films were incubated overnight in each solution (11 mM), mentioned in Table 2 followed by 24 h incubation in phage solution (2ml) at 37ºC. The phage-functionalized films (S1-S5, S7) were washed thrice with sterile water to remove any unbound phages and then tested for their lytic efficacy against, SDKU6, using plaque assay. The CTAB modified PVDF membrane (sample S6) was taken as positive control, which is reported to have bactericidal activity, whereas PVDF alone (Sample S8) was used as negative control for infectivity study (Gilbert and Al-taae, 1985).
Results
Isolation and Characterisation of Bacteria
Six of the isolated bacteria, viz. SDKU, SDKU2, SDKU3, SDKU4, SDKU5 and SDKU6 were characterized individually by Gram staining and biochemical tests (Table 3) (Prauser et al., 1985). One of the bacterial isolates, SDKU6 showed positive results with respect to mannitol salt agar, nitrate reduction, catalase and glucose fermentation. It was further characterized in detail by ribotyping, which generated a restriction pattern from 16S rRNA gene that was compared with available standard database on bacterial genus and species and identified as Staphylococcus arlettae (Altschul et al., 1990; Stackebrandt and Goodfellow, 1991). Table 3 presents the results obtained from morphological, biochemical and phylogentic analysis of SDKU6.
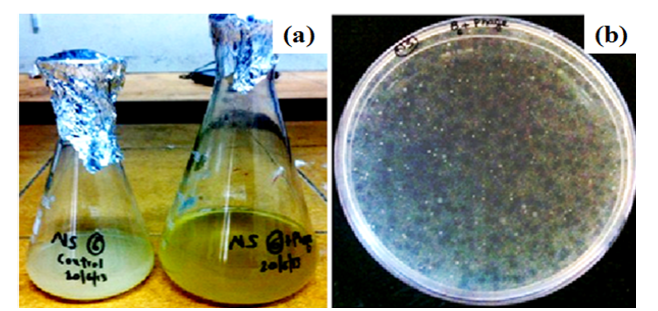
Figure 3: Isolation of phages against SDKU6 by (a) Tubidity disappearance and (b) Agar overlay Method
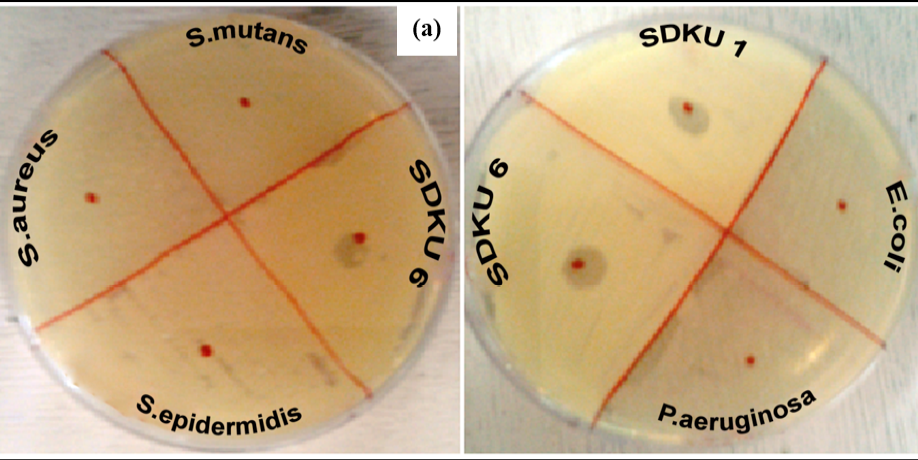
Figure 5: Host specificty test of phages against (a) Gram positive and (b) Gram negative bacterial strains
The results of antibiotic sensitivity tests of bacteria (Staphylococcus arlettae SDKU and Staphylococcus arlettae SDKU6) are given in Table 4 in the form of zones of inhibition and interpretation of susceptibility was obtained by comparing the results to the standard zone sizes using the criteria published by the Clinical and Laboratory Standards Institute (CLSI, formerly the National Committee for Clinical Laboratory Standards or NCCLS). Figure 2 shows the results of antibiotic susceptibility test in graphical form.
Isolation and Characterisation of Bacteriophages
Figure 3 is associated with the characterization of phages isolated against SDKU6 from sewage water. For enrichment of phages, the sample filtrate was incubated overnight with SDKU6 broth culture (section 2.1.2) after which the turbidity of medium containing water sample disappeared while control (without water sample) remained turbid due to bacterial growth (Figure 3a). The bacteriophages against SDKU6 were extracted from enriched phage solution by double agar overlay method. Figure 3b shows that a large number of clear zones (plaque) of diameter ~2 mm appeared on agar plate, thus establishing the presence of lytic phages in enriched broth. The titre of purified phage solution was found to be 5.8 ×108 pfu/ml, which was used further for immobilization studies.
Table 4: Measured zones of inhibition of bacteria for susceptibility testing to antibiotics
|
Charaterisation |
Test |
Result |
|
Morphological |
Gram Staining |
Purple colored, cocci shaped |
|
Biochemical Tests |
Mannitol Salt Agar |
Positive |
|
Catalase test |
Positive |
|
|
Nitrate reductase test |
Positive |
|
|
Glucose ferementation |
Positive |
|
|
Phylogenetic |
16S rRNA Sequencing |
GenBank KF589201 |
The corresponding morphology of isolated and purified phages was determined by TEM (Figure 4), showing a large number of phages arranged in clustered manner. The phages possess an icosahedral head of width ~ 60 nm, and a long non-contractile tail of length ~150 nm with tail fibres (Figure 4) .
Figure 5a shows lytic activity of phages against bacterial strain SDKU6, while no activity was observed against other gram-positive bacteria, namely, S. aureus, S. epidermitis, and S. mutans. Similarly, when tested against gram negative bacteria E. coli and P. aeruginosa, no lysis zones appeared as evident from Figure 5b.
Synthesis and LB Film Formation of Silica NPs
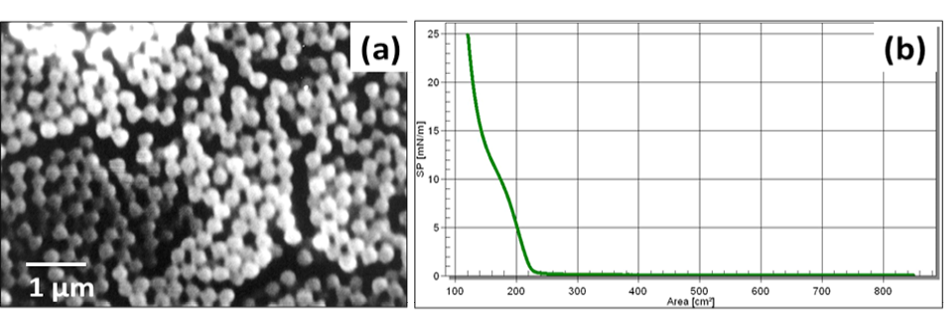
Figure 6a presents SEM image of synthesized silica NPs of size ~ 300 nm. Figure 6b is associated with the isotherm behavior of the synthesised silica NPs at air/water interface.
Immobilization of Phages onto Silica NPs LB Film
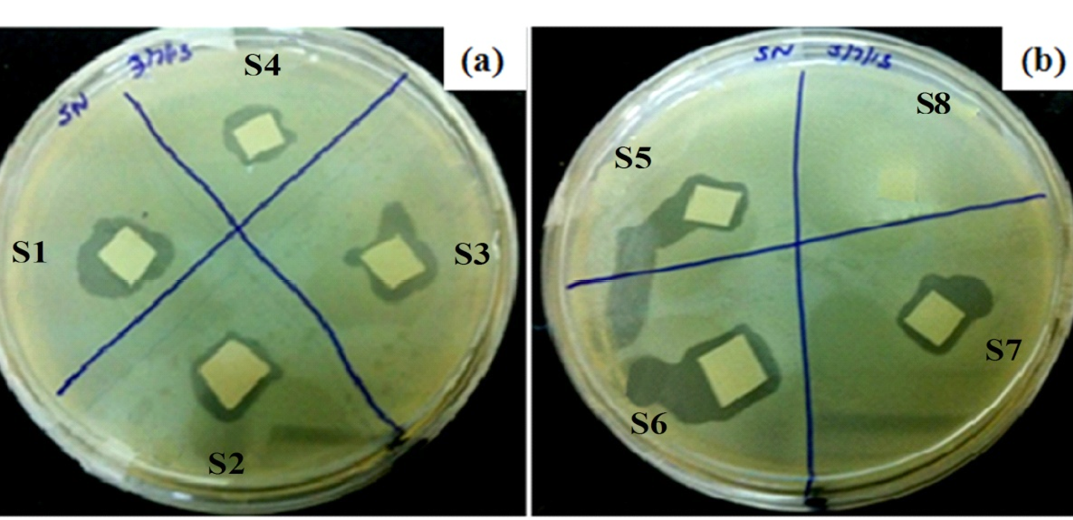
Figure 7 corresponds to the lytic efficiency of each phage-functionalized platform, S1 to S8 (Table 1). It can be seen that no zone of lysis appeared in negative control (S8) i.e. PVDF membrane without phages, whereas lysis ring around membrane was observed with all other samples, including positive control S6. Functionalized films S1, S4 and S2, S3 showed prominent clear zones in comparison to S5 and S7, where phages were bound to membrane via physisorption.
Discussion
Isolation and Characterisation of Bacteria
Six morphologically different bacterial colonies, observed on the agar plates were characterized using Gram staining and biochemical tests. Gram staining differentiates bacterial species largely into two groups namely, Gram- positive and Gram- negative, based on chemical and physical properties of their cell walls (James et al., 2013). All six of the isolated bacterial strains revealed purple colored and cocci shaped microbial cells upon Gram staining under compound microscope, which is a characteristic of Gram-positive bacteria. Two of the isolated strains, SDKU6 and SDKU, showed positive test with mannitol salt agar, which is reported to be a selective and differential medium specifically for Staphylococcus bacterial species. The results of other biochemical tests such as catalase, nitrate-reductase, glucose fermentation were in agreement with above hypothesis, which manifests that the SDKU and SDKU6 are of Staphylococcus genus (Prescott et al., 2002b). 16S rRNA ribotyping and comparison with database using BLAST of SDKU6, showed 99% similarity to Staphylococcus arlettae bacterial strain, which was further verified by phenotypic analysis reports provided by IMTECH, Chandigarh, India (Altschul et al., 1990; Stackebrandt and Goodfellow, 1991). Additionaly, a unique accession number, KF589201, has been assigned by GenBank to the isolate. SDKU was also identified as Staphylococcus arlettae by IMTECH, Chandigarh, however, it exhibits different growth conditions, colored pigment in comparison to SDKU6, which can be attributed to the subspecies of S. arlettae (Source: IMTECH report). It can be said from the results of antibiotic susceptibility tests (Figure 4 and Figure 2) that the bacteria Staphylococcus arlettae SDKU and SDKU6 are completely resistant to Methicillin and Amphotericin.
Isolation and Characterisation of Bacteriophages
The bacteriophages were isolated from sewage water against one of the foot odour causing bacterial isolates, i.e. SDKU6. The clear solution obtained after enrichment (Figure 3a) shows the presence of phages against SDKU6 bacterium as without phages, the solution became turbid due to the growth of bacteria upon incubation at 37ºC for 24h (control). Further, to isolate phages from enriched solution, double agar overlay method was employed. The appearance of large number of plaques (clear zones) onto agar plate (Figure 3b) confirmed the presence of phages in enriched sample filtrate. The phage pellet obtained after ultracentrifugation was redispersed in PBS buffer (5ml) resulting in high titre purified phage stock solution. The purified phages were characterized partially by determining their host specificty and morphology. Since electron microscopy is reported as one of the fastest and easiest ways of phage identification other than by sequence analysis to assign phages to a family, morphological analysis of isolated phages was done using TEM (Figure 4) (Kutter and Sulkvelidze, 2005). The phages possess a long, non-contractile tail alongwith an elongated head (as evident from Figure 4), therefore they can be attributed to Caudovirales order and Siphoviridae family of phages (Synott et al., 2009). From the study of host specificity (Figure 5), it was found that phages, isolated against SDKU6, did not infect any other Gram positive and Gram negative bacterial species tested in this study except SDKU6 and SDKU. It was interesting to note that the phages isolated against SDKU6 also lysed SDKU bacteria, as it was also identified as S. arlettae species (from IMTECH laboratory), whereas, the reverse case was not true (Data not shown). The absence of clear zone against other bacterial strains used in our study revealed that their host range is limited only to Staphylococcus arlettae species of bacteria. This specificity of interaction of a bacteriophage with its host is advantageous for their usage in control of bacteria of a particular species (Armon and Schwartz, 2009; Banowski and Simmering, 2011; Pearson et al., 2013; Shin et al., 2011; Yang et al., 2013).
Synthesis and LB Film Formation of Silica NPs
Since silica NPs offer advantages such as low cost of synthesis, high surface area and easy modification, these were synthesised and used in the study (He and Hu, 2004). Figure 6a shows the morphology of synthesized silica NPs and reveals that the particles are highly monodispersed and spherical in nature. The size of the particle is dependent upon several parameters such as precursor concentration, type and amount of alcohol, reaction time as well as temperature (Devi et al., 2013). The synthesised NPs were deposited over a PVDF membrane using LB technique in the form of a monolayer at solid phase. The monolayer formed can be observed from the isotherm (Figure 6b) which manifests three distinct phases, III, II and I corresponding to solid, liquid and gas phases, respectively.
Immobilization of Phages onto Silica NPs LB Film
Six approaches were used for phage immobilization, based on covalent, electrosatic and physiosorption interactions, listed in Table 2 (schematic shown in Figure 1). The clear zones associated wirh the lytic efficiency of phage bound membranes is shown in Figure 7, which concludes that the phages retained their infectivity even after immobilization. The APTMS and AEPTMS are known to have positively charged amine functionalities, which were used to electrostatically bind phages through the negatively charged carboxyl groups present on their head in samples S1 and S4. In contrast, gluteraldehyde, which is a homobifunctional cross linker, was used to bind phages covalently on modified silica NPs LB film. It can also be observed from Fig. 7 that the phages immobilized on silica NPs LB film after modification with APTMS and AEPTMS (S1 and S4) show a very uniform and wide lytic ring due to strong electrostatic interaction between cationic film surface having amine groups and anionic phage heads. Similarly, wide and uniform lytic zones were also observed with covalent linkage using gluteraldehyde as a cross linker (S2 and S3). It can be said from Figure 7 that the phages anchored to appropriately modified surfaces in proper orientation with their head attached to a surface and tails kept free and thus, have high infectivity and capture efficiency (Anany et al., 2011). Also, the effect of NPs high surface/volume ratio property can be observed by seeing prominent lysis zone (sample S5), in comparison to S7, where PVDF was used directly for immobilization by physical adsorption. This is because high aspect ratio of NPs enables high biomolecular loading (Bashar et al., 2013; Hutter and Maysinger, 2013; Wang et al., 2008).
Thus, the present work is an intial attempt to pave a path for use of phages as a prominent antibacterial preparation to prevent Methicillin-resistant bacerial infections in animals and humans. The approach can be extended further for development of antibacterial dressings using a biodegradable polymer matrix impregnated with a cocktail of bacteriophages having lytic activity against multidrug resistant bacteria. However, further study is needed for evaluation of the associated lytic efficiency of phage immobilised surfaces with time.
Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgements
Authors are thankful to Council of Scientific and Industrial Research, India and University Institute of Engineering and Technology, Kurukshetra, India for providing infrastructural facilities. The authors also declare that they have no conflict of interest.
References