Advances in Animal and Veterinary Sciences
Research Article
Advances in Animal and Veterinary Sciences 2 (6): 321 – 328InVitro Characterization of Listeria monocytogenes Isolates by Haemolysis, Camp, Piplc Assay with Protein Profiling and Antibiotic Resistance Recovered from Nagpur Region
Nitin Chandhrakant Dudhe1*, Sandip Prabhakar Chaudhary1, Pranita Bhanudas Pantawane2, Vinod Namdeorao Hirde1, Thadiyam Puram Ramees3, Nandkishor Namdeorao Zade1
- Department of Veterinary Public Health & Epidemiology, Nagpur Veterinary College, Seminary hills, Nagpur 440006, India
- Department of Veterinary Public Health, Bombay Veterinary College, Parel, Mumbai, 400012, India
- Division of Veterinary Public Health, Indian Veterinary Research Institute, Izatnagar, Bareilly (U.P.) – 243122, India
*Corresponding author: drnitindudhe86@gmail.com
ARTICLE CITATION:
Dudhe NC, Chaudhary SP, Pantawane PB, Hirde VN, Ramees TP, Zade NN (2014). InVitro characterization of Listeria monocytogenes isolates by haemolysis, CAMP, PIPLC assay with protein profiling and antibiotic resistance recovered from Nagpur region. Adv. Anim. Vet. Sci. 2 (6): 321 – 328.
Received: 2014–05–26, Revised: 2014–07–03, Accepted: 2014–07–04
The electronic version of this article is the complete one and can be found online at
(
http://dx.doi.org/10.14737/journal.aavs/2014/2.6.321.328
)
which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
ABSTRACT
Listeria monocytogenes is one of the most virulent foodborne zoonotic pathogen with 20 to 30% of clinical infections resulting in death. The L. monocytogenes is ubiquitous in the nature and has been isolated from a variety of foods and environmental sources infecting human and animals revealing the pathogenicity of organism. Reports are very few regarding the pathogenesis of organism in Nagpur region, Maharashtra, India. So the present study was proposed to evaluate the pathogenicity of 12 L.monocytogenes isolates recovered from foods of animal origin (Chevon, pork, beef and carabeef) collected in and around Nagpur city. In vitro expression of pathogenisity was carried out by using viz., Haemolysis on Sheep blood agar (SBA), Christie Atkins Munch Petersen (CAMP) Test, Phosphatidylinositol–specific phospholipase–C (PI– PLC) assay along with protein profiling and subsequently studied for their pattern of antibiotic resistance. All 12 isolates of L. monocytogenes were found haemolytic in nature and showed PIPLC activity with variation in degree of production which designated them as pathogenic one. The protein profiles of all isolates exhibited minimum 25 to maximum 29 bands where protein bands of molecular weight from 56.237 kDa to 60.33 kDa and 29.226 kDa to 33.48 kDa shared by all isolates can be correlated with the haemolytic LLO (Listerolysin–O) and PI–PLC activity respectively. In antibiotic assay, most isolates showed multidrug resistance, besides no resistance was recorded towards Gentamycin, Norfloxacin, Ampicilin and Cefotaxime. The study revealed a high virulence nature of isolates of L. monocytogenes among food animals in and around Nagpur region. Haemolysin on SBA, PI–PLC activity along with exhibition of polypeptide in the range for these activities can be suggestive of protein profiling as technique for determination of pathogenicity
INTRODUCTION
Surveillance of food borne diseases is utmost priority in the public health agenda worldwide, as a leading cause of death from a food borne pathogen, Listeria monocytogenes continues to cause sporadic cases and outbreaks of illness. The organism is one of the most virulent food borne zoonotic pathogen having very high mortality rate (Schlech, 2010) and the reports are increased in prevailing years worldwide (EFSA, 2012). Recently major outbreaks are reported in American and European countries (Todd and Notermans, 2011), revealing to date huge public health significance. The organism is ubiquitous in nature and widespread in environment inevitably results in contamination of numerous food products. Many attempts has been made to isolate Listeria spp. from a variety of foods and environmental sources such as water, sludge, soil, plants, vegetation, food, infecting humans and animals (Liu, 2008; Dhama et al., 2013). However, the published information on the status of pathogenecity of L. monocytogenes from these foods is very scattered and unsystematic, both in the veterinary and public health sectors (Khan et al., 2013). Till date the cases of abortion, stillbirths, neonatal septicemia and encephalitis have been documented in human beings and domestic animals (Jamali and Radmehr, 2013).
To assess the virulence properties of a particular L. monocytogenes isolate, the mouse pathogenicity and chick embryo test were preferred, for this skilled personnel and animal ethics are major concerns. Therefore alternative methods ideally should be based on the phenotypic characterization by in–vitro methods as Haemolysis, CAMP and PIPLC assay
Hemolysis is an important characteristic which would seem to be directly related to the pathogenicity of Listeria, since non–haemolytic Listeria species can be considered as non–pathogenic (FAO/WHO/OIE, 2008) and the pathogenicity of the pathogen is highly correlated to the haemolytic factor (Yadav et al., 2010; Momtaz and Yadollahi, 2013). Considering haemolysis, only L. monocytogenes and L. ivanovii produced characteristic haemolysin like listeriolysin–O (LLO) and ivanolysin–O, respectively.
In CAMP test, haemolysin acts synergistically with the β– haemolysis of S. aureus on sheep erythrocytes and gives synergistic zone of hemolysis towards S. aureus which is due to either phosphatidylinositol specific or phosphatidyl–choline specific C from L. monocytogenes and sphingomyelinase from Staphylococcus aureus (Farber and Peterkin, 1991; McKellar, 1994). Evaluation of the pathogenicity of Listeria species by phosphatidylinositol specific phospholipase C (PI– PLC) based assay has been reported to be reliable indicator to discriminate pathogenic and non–pathogenic Listeria species (Notermans et al., 1991). Copious workers have been used CAMP test to charecterised L. monocytogenes isolates (Tasci et al., 2010; Mahmoodi, 2010; Gebretsadik et al., 2011). In recent years, many resistant strains have been detected of listeriosis (Charpentier and Courvalin, 1999) this may be due to indiscriminate use of antibiotics results in the inflation of multi–drug resistance in L. monocytogenes bacteria, so antibiogram studies can be very useful in deciding line of treatment.
Keeping things in mind, the published information on status of L. monocytogenes in meat and methods available for the detection of pathogenic L. monocytogenes, we investigated the pathogenecity of L. monocytogenes isolates recovered from various meat samples in Nagpur, India, by examining in vitro characterization of L. monocytogenes isolates via Haemolytic, CAMP and PIPLC assay along with protein profiles.
MATERIALS AND METHODS
Isolates
A total of 12 isolates of Listeria monocytogenes were isolated from meat samples and maintained in Division of Veterinary Public Health, Nagpur veterinary college, Nagpur Maharashtra and used for expression of in–vitro pathogenicity, protein profiling and antibiotic sensitivity tests.
Biologicals
The standard bacterial strains of Listeria monocytogenes (MTCC 1143), Staphylococcus aureus (MTCC 3160) and Rhodococcus equi (MTCC 1135) were obtained from Microbial Type Culture Collection (MTCC) and Gene Bank, Institute of Microbial Technology, Chandigarh, India. Chemicals/conjugates required for the study were procured from Sigma Aldrich (US) and Merk/Gene bank, India. The organisms were maintained on brain heart infusion agar slopes at 4oC and used for preparation of LLO.
Haemolysis on Sheep Blood Agar (SBA)
Detection of haemolysin production was performed on sheep blood agar (SBA) as per Cruickshank et al., (1975). Freshly grown isolates were streaked onto blood agar plates prepared using defibrinated sheep blood (5 per cent), incubated at 370C for 24 hrs and observed for the zone of haemolysis (Courtieu, 1991).
Christie Atkins Munch Petersen (CAMP) Test
The isolates were subjected to CAMP test as per the method of ISO (1996). Isolates of Staphylococcus aureus (MTCC 3160) and Rhodococcus equi (MTCC 1135) were freshly grown in Brain Heart Infusion (BHI) broth for 18–24 hrs and streaked onto freshly prepared SBA plates (7 %) in a manner of straight line with a distance of 3–4cm so that the streaks were wide apart and parallel to each other. Freshly grown isolates of L. monocytogenes were streaked in between two parallel streaks of S. aureus and R. equi at angle of 900 leaving a space of three mm against parallel streaks. The plates were incubated at 370C for 24 hrs and observed for enhanced zone from partial hemolysis to a wider zone of complete hemolysis.
Phosphatidylinositol–Specific Phospholipase–C (PI– PLC) Assay
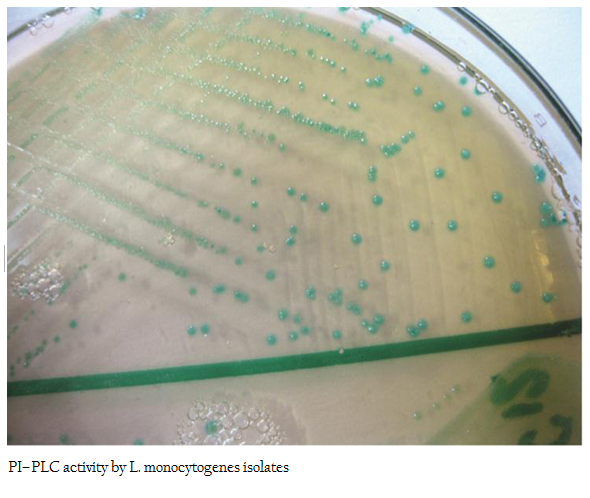
The PI– PLC assay was performed according to the standard procedure described by Yadav et al. (2010). The isolates of Listeria were overgrown on SBA 5 per cent Figures at 370C. These isolates were freshly grown in BHI broth for 18–24 hrs and then streaked onto L. mono differential agar (Hi Media Ltd, Mumbai, India). The inoculated Figures were incubated at 370C in a humidified chamber for 24 hrs. Development of light blue colonies were considered positive for PI–PLC assay and accordingly designated as pathogenic.
Protein Profile Employing SDS– PAGE
The protein profiling studies of L. monocytogenes isolates was employed as per the standard procedure described by Lammeli, (1970). All isolates of L. monocytogenes were inoculated in 20 ml BHI broth with continuous aeration (shaker) at 370C for 12 hrs. The grown cultures were then centrifuged at 40C at 10,000 rpm for 5 min (REMI, India) to harvest the supernatants. The culture supernatants were then filtered through nitrocellulose acetate filters (0.45μ) to cell free culture supernatants. The cell free culture supernatants were processed for protein estimation as per Lowry et al., (1951) and 24 μl of each were loaded onto lane of SDS–PAGE.
Sodium–Dodecyl Sulphate Polyacrylamide Gel Electrophoresis (SDS–PAGE)
In order to determine the molecular weight of purified protein, vertical polyacrylamide slab gel (12.5 %) and stacking gel (5 %) was casted in assembly (Banglore, Genei). An amount of approximately containing 7 μg proteins was mixed in 5x sample buffer. The mixture was boiled for three minutes and loaded after cooling in lane of each gel. Assembly was run at 40V (till stacking gel) and further at 90V (in separating gel). The dye front was noted for migration and completion of run. The protein molecular weight marker of medium range (Banglore, Genei) comprising of 97.4 kDa, 66 kDa, 43 kDa, 29 kDa, 20.1 kDa and 14.3 kDa was used as standard. The protein profiles were visualized by employing silver staining technique as per Rosenberg, (1996). The gels were photographed using gel documentation system (BioRad, US) and further calculations of molecular weights (MW) of the peptides were done by extrapolation of relative mobility of the unknown protein against that of standard molecular weight marker using quality– One software (BioRad, US).
Antibiotic Sensitivity Test
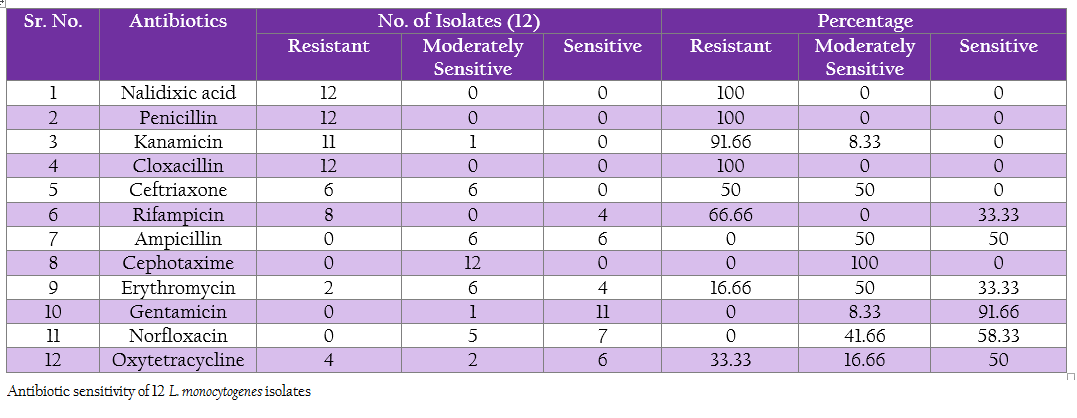
Antibiotic sensitivity of L. monocytogenes isolates to various antibiotics and chemotherapeutic agents was studied by agar disc diffusion method using single antibiotic disc (Bauer et al., 1966). The selection of antibiotic was based on the routinely used antibiotic in field viz., Ampicillin, Cephotaxime, Cloxacillin, Erythromycin, Gentamicin, Kanamycin, Nalidixic acid, Norfloxacin, Penicillin–G, Rifampicin, Oxytetracycline and Ceftriaxone (Table 12).
The isolates were freshly grown in BHI broth for 18–24 hrs and then spread evenly on BHI agar plates by using sterile cotton swabs. The antibiotic discs were then placed with the help of sterile forceps and plates were incubated at 37oC for 24 hrs. After incubation, zone of inhibition around each antibiotic disc was measured and each isolate was noted as a sensitive, moderately sensitive and resistant against respective antibiotic based on the size of inhibition zone according to manufactures instruction (Hi–Media Mumbai).
RESULTS
All 12 isolates of L. monocytogenes were found haemolytic with variation in degree of haemolysin production based on which differentiated into strong and weak haemolysin producing strain. However among 12 isolates, isolate p49 showed extensive haemolytic zone while remaining 11 isolates showed a mild zone of haemolysis (Figure1; Table 1). Significant enhanced zone of haemolysis was produced by all 12 isolates of L. monocytogenes towards S. aureus (MTCC 3160) and subsequently no zone was recorded towards R. equi (MTCC1143). Similarly, the isolates showed cent percent positivity in case of PIPLC activity. However, the results are varied as strong PI–PLC producer (++) in case of isolates from cattle (C14) and other for goats (G21) were noted (Figure 2 and 3; Table 1).
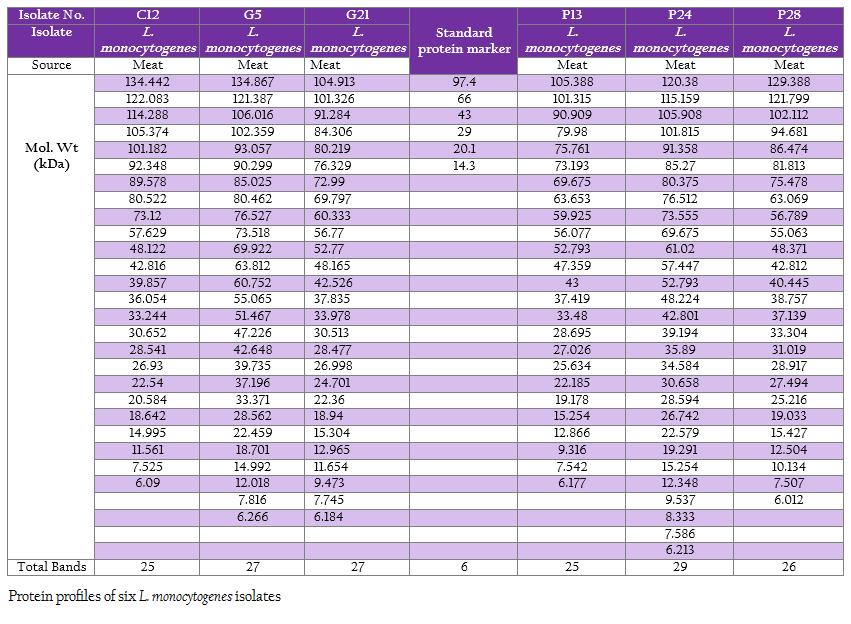
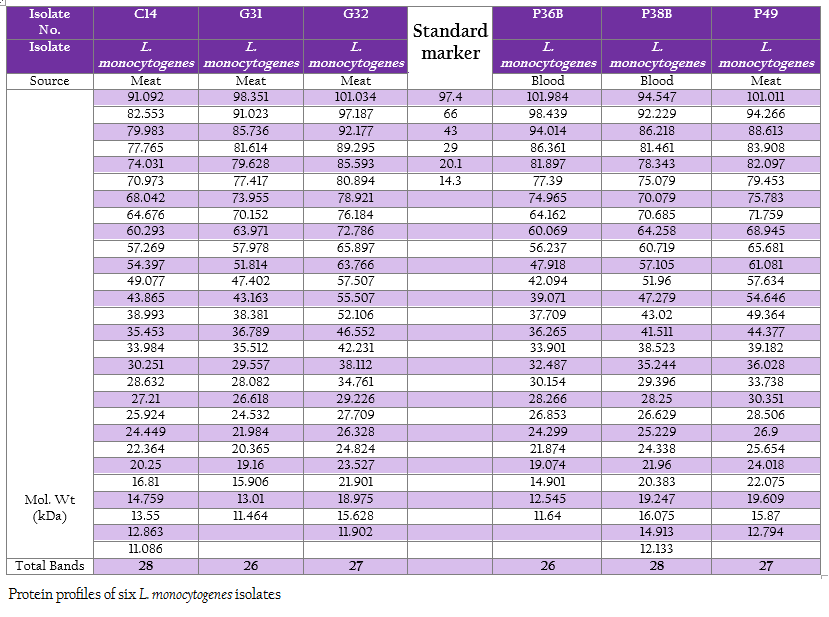
The protein profiles of all isolates exhibited minimum 25 to maximum 29 bands ranging from lowest 6.090 kDa to 134.867 kDa (Figure 4a and 4b). The presence of protein bands of molecular weight from 56.237 kDa to 60.33 kDa shared by all isolates of L. monocytogenes (Table 2a and 2b) can be representing haemolysin LLO. Parallelly these isolates were found haemolytic on SBA and thereby the ability of isolate to invade the host cell confirming its pathogenicity. The isolates also exhibited and shared polypeptide in the range of 29.226 kDa to 33.48 kDa (Table 2a and 2b). In antibiotic assay, all isolates of L. monocytogenes (100%) were found to be resistant to one or more antibiotics. Nalidixic acid, Penicilin and Cloxacillin exhibited 100% resistance followed by Kanamycin, Rifampicin, Ceftriaxone, Tetracycline and Erythromycin
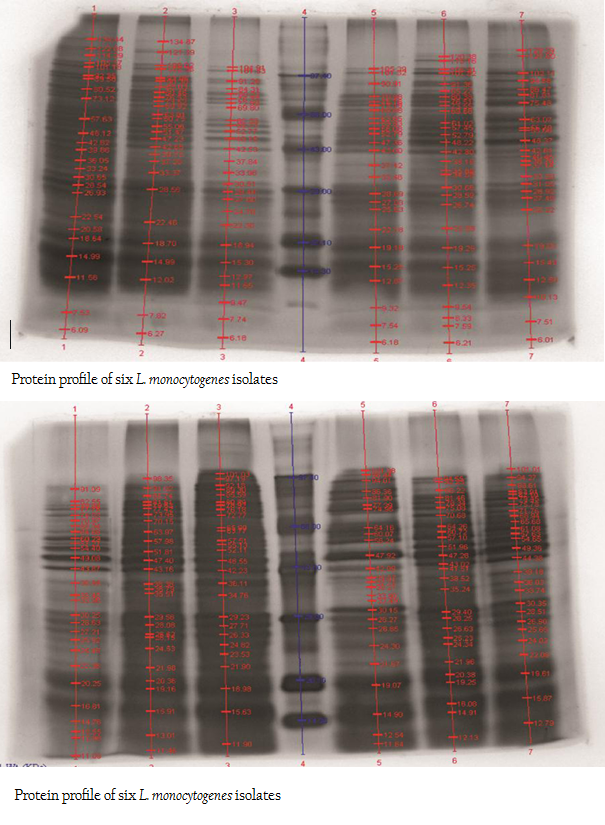
Figure 4: A: Protein profile of six L. monocytogenes isolates; B: Protein profile of six L. monocytogenes isolates
was in 91.66, 66.66, 50, 33.33 and 16.66% of strain respectively. No resistance was recorded towards Gentamycin, Norfloxacin, Ampicilin and Cefotaxime. Highest sensitivity of 91.66% was found against Gentamycin (Table 3).
DISCUSSION
Multiple virulence factors such as hemolysin, CAMP, PIPLC assay are important in the pathogenesis of L. monocytogenes infections (Rawool et al., 2007) and indicate the potential of virulent strains to invade host cells and cause listeriosis (Ryser and Marth, 2007).
The L. monocytogenes phospholipases are vital determinants of pathogenicity (Marquis et al., 1995) expressed solely by pathogenic spp. of Listeria which is haemolytic i.e., L. monocytogenes (Liu, 2004). For that reason, these assays remain a reliable indicator to demonstrate and differentiate the pathogenicity of L. monocytogenes (Notermans et al., 1991).
β–haemolytic activity of L. monocytogenes isolates is associated with putative CAMP like factor (Koprnspan et al., 2014). The cent percent haemolytic activity showed by L. monocytogenes isolates has been reported by several researchers (Bhanu Rekha et al., 2006; Molla et al., 2004; Yucel et al., 2004; Osman et al., 2014) the same as in the present investigation and those isolates were further designated as the pathogenic one (Barbuddhe et al., 2000, Lacier and Centorbi, 2002; Chaudhari et al., 2004; Becker et al., 2006; Bhanu Rekha et al., 2006; Jallewar et al., 2007; Elezebeth et al., 2007) reveals the strong activity of LLO. Variation in haemolytic activity attributed to composition of medium, and/or a loss of ability to produce hemolysin may have occurred over successive passages through culture media which impaired the identification of these cultures as L. monocytogenes (Nunes and Hofer, 1994).
CAMP test is a diagnostic tool that reliably and quickly provides presumptive identification of L. monocytogenes (Savini et al., 2014). In CAMP assay, enhanced zone of haemolysis showed by all 12 isolates towards S. aureus designates them as pathogenic one seeing that synergistic effect of Listeriolysin–O (LLO) and Streptolysin–O (SLO) has directly implicated the virulence of LLO (Buchanan et al., 1989; McKellar, 1994; Barbuddhe et al., 2000, Molla et al., 2004; Becker et al., 2006; Bhanu Rekha et al., 2006; Abd EI–MaIek et al., 2010; Nayak et al., 2010; Gebretsadik et al., 2011). Further cent per cent PI–PLC activity of all 12 isolates adds to the pathogenic potential (Paziak–Domanska et al., 1999; Yadav et al., 2010; Momtaz and Yadollahi, 2013) consequently it is acted as a one of the best characterized virulence factor (Moreno et al., 2014; Park et al., 2014).
Variation in haemolysis and PIPLC activities was observed in study as one isolate (p49) found to be extensive haemolytic and others remains a mild haemolytic, similarly in PIPLC assay two isolates showed a strong acitivities (C14 and G21) while others showed mild activities, the disparity in the performance might be due no coordination of genes encoding for these activities or lack of virulence determinants due to mutation in the strain of L. monocytogenes (Notermans et al., 1991; Shakuntala et al., 2006; Kaur et al., 2010). Overall, in the present investigation all (12) isolates of L. monocytogenes isolated from food animals turn pathogenic in all tests employed, indicating their high virulence activity. To date, CAMP remains a reliable indicator for diagnosis of pathogenesity of organism (Savini et al., 2013) and may be performed directly to exclude or confirm L. monocytogenes (Bhat et al., 2013; Savini et al., 2014).
SDS–extracted patterns in protein profiling are strictly specific for species except a few major bands appear to be common to all listeria species (Tabouret et al., 1992). The presence of protein bands from 56.237 kDa to 60.33 kDa and consequently haemolytic activity indicated the production of β–Listeriolysin (55 – 60 kDa), which are also probably responsible for the positive CAMP phenomenon and this protein band (50–60 kDa) may eventually emerge as significant factor of Listerial pathogenicity as described by Parrisius et al. (1986). Also, all isolates exhibited and shared polypeptide in the range of 29.226 kDa to 33.48 kDa which can be representing PI–PLC as reported by Notermans et al. (1991) since these isolates were showed PI–PLC activity on L. Mono differentiating agar.
In present study, 100% resistance was showed against Nalidixic acid and Penicilin, the results are in accordance with Yucel et al. (2005); Ennaji et al. (2008); Ruiz–Bolivar et al. (2011) and Gunjal (2006); Pozark and Murano (2002) respectively. Whereas, Rahimi et al. (2010) recorded 96.4 % resistance for the Penicilin and Lotfolahi et al. (2011) revealed 77.77% of resistance towards Penicilin. Similarly, Kanamycin showed 91.66% resistance which has also noticed by Ruiz–Bolivar et al. (2011). In contrast, Rodas–Suaerez et al. (2006); Nwachuwu et al. (2010) and Morvan et al. (2010) reported 22.5, 40.91 and zero percent resistant for penicillin respectively. Nwachukwu et al. (2010) reported 22.22% resistance for Rifampicin while in present study 66.66%. In case of Tetracycline, 16.66% resistant was noticed which varies from the result of Rodas–Suarez et al. (2006) and Gunjal (2006) who reported 2.3 and 8.16 % resistance respectively. Variation in the geographical region and differences in therapeutic practices can think to be major contributing factors.
Overall 91.66 per cent of sensitivity was observed for Gentamicin, the same was observed by Wong et al. (1990), Walsh et al. (2001), Ennaji Hayat et al. (2008), Morobe et al. (2009) and Vasu et al. (2014) as 99.4, 99, 100, 84.21and 100% , respectively. These deviations in the antibiotic resistance pattern attributed to the fact that antibiotic resistance is encoded by the plasmid which has capability to transfer from one strain to another (Mayer, 1988). However, the presence of multi–drug resistance among virulent strains of L. monocytogenes in the region is pointing to increase in potential threat from public health point of view (Prazak et al., 2002).
CONCLUSION
The observation on exhibition of polypeptide in the range of 50–60 kDa indicative of LLO and its parallel expression as production of haemolysin on SBA as well as exhibition of protein band in the range of 29–33 kDa along with in–vitro demonstration of PI–PLC activity on agar can be suggestive of protein profiling as technique for determination of pathogenicity. Bearing of the proteins with common molecular weight among these isolates as well as L. monocytogenes isolates from the earlier studies can be exploited for study of epidemiological marker for the region specific isolates. Further, large number of samples will be needed to draw any conclusion pertaining to ‘conserved protein profile’ specific to the region. However, subsequent exhaustive studies are required in this direction to build up the remark on this matter.
ACKNOWLEDGEMENTS
Authors are thankful to department of Veterinary Public Health, Nagpur Veterinary College Nagpur, Maharashtra, India for providing all necessary facilities to carried out research work.
REFERENCES
Abd El–Malek A, Hassan SF, Hassanein R, Abdelaxzeem M, Mohaned, Elsayh KJ (2010). Occurrence of Listeriaspecies in meat, chicken products and human stools in Assiut city, Egypt with PCR use for rapid identification of Listeria monocytogenes. J. Vet. World. 3(8): 353 – 359.
Barbuddhe SB, Malik SVS, Bhilegaonkar KN, Kumar P, Gupta LK (2000). Isolation of Listeria monocytogenesand anti–listeriolysin–O detection in sheep and goats. Small. Rumi. Res. 38: 151 – 155.
http://dx.doi.org/10.1016/S0921-4488(00)00155-3
Bauer AW, Kirby MN, Sherris JC, Truck M (1966). Antibiotic susceptibility testing by standardised single disc method. Amer. J. Clin. Pathol. 45: 443 – 446.
Becker B, Schuler S, Lohnesis M, Sabrowiski A, Gordon DW, Curtis W, Holzapfel H (2006). Comparison of two chromogenic media for the detection of L. monocytogenes with the plating media recommended by EN/DIN 11290–1. Intl. J. Food Microbiol. 109: 127 – 131.
http://dx.doi.org/10.1016/j.ijfoodmicro.2006.01.030
PMid:16515816
BhanuRekha V, Malik SVS, Chaudhari SP, Barbuddhe SB (2006). Listeriolysin O–based diagnosis of Listeria monocytogenesinfection in experimentally and naturally infected goats. Small Rumi. Res. 66: 70 – 75.
http://dx.doi.org/10.1016/j.smallrumres.2005.07.021
Bhat SA, Willayat MM, Roy SS, Bhat MA, Shah SN, Ahmad A, Maqbool, Ganayi BA (2013). Isolation, molecular detection and antibiogram of Listeria monocytogenes from human clinical cases and fish of Kashmir, India. Comp. Clin. Pathol. 22(4): 661 – 665.
http://dx.doi.org/10.1007/s00580-012-1462-1
Buchanan RL, Heidi, Stahl G, Marianne, Bencivengo M, Corral FD (1989). Comparison of Lithium Chloride–Phenylethanol–Moxalactam and Modified Vogel Johnson Agars for detection of Listeria spp. in retail–level meats poultry and seafood. Appl. Environ. Microbiol. 55: 599 – 603.
PMid:2494936 PMCid:PMC184166
Carpentier E, Courvalin P (1999). Antibiotic resistance in Listeria spp. Antimicrob. Agents Chemother. 43(9): 2103 – 2108.
Chaudhari SP, Malik SVS, Barbuddhe SB (2004). Humoral and delayed type hypersensitive responses against Listeria monocytogenes phosphatidylinositol–specific phospholipases C in experimentally infected buffaloes. Vet. Res. Comm. 28: 569 – 579.
http://dx.doi.org/10.1023/B:VERC.0000042864.98892.06
PMid:15563104
Courtieu AL (1991). Latest news on Listeriosis. Comp. Immun. Microbiol. Infect. Dis. 14: 1 – 7.
http://dx.doi.org/10.1016/0147-9571(91)90035-C
Cruickshank R, Dugaid JP, Marmion BP, Swain RHA (1975) Med. Microbiol. 2: 587.
Dhama K, Verma AK, Rajagunalan S, Kumar A, Tiwari R, Chakraborty S, Kumar R (2013). Listeria monocytogenes infection in poultry and its public health importance with special reference to food borne zoonoses. Pak. J. Biol. Sci. 16(7): 301–308.
http://dx.doi.org/10.3923/pjbs.2013.301.308
PMid:24498796
Elezebeth G, Malik SVS, Chaudhari SP, Barbudhe SB (2007). The occurance of Listeria spp. and antibodies against listeriolysin–O in naturally infected goats. Small Rumin. Res. 67: 173 – 178.
http://dx.doi.org/10.1016/j.smallrumres.2005.09.029
Ennaji H, Timinouni M, Ennaji M, Hassar M, Cohen N (2008). Characterization and antibiotic susceptibility of Listeria monocytogenes isolated from poultry and red meat in Morocco. Infect. Drug Resis. 1: 45 – 50.
European Food Safety Authority (2012). European Food Safety Authority "Schmallenberg virus": analysis of the epidemiological data and assessment of impact assessment. EFSA J. 10(6): 2768.
FAO/WHO/OIE (2008). Joint FAO/WHO/OIE Expert meeting on critically important antimicrobials. In: Report of a meeting held in FAO, Rome, Italy, 26–30November 2007, FAO, Rome, Italy and WHO, Geneva, Switzerland.
Farber JM, Peterkin PI (1991) Listeria monocytogenes, a foodborne pathogen. Microbiol. 55: 476 – 511.
Gebretsadik S, Kassa T, Alenaychu H, Huruy K, Kebede N (2011). Isolation and characterization of Listeria monocytogenes and other Listeria species in foods of animal origin in Addis Ababa, Ethiopia. J. Inf. Pub. Hlth. 4: 22 – 29.
Gunjal PS (2006) Genotype characterization of Listeria spp. from raw poultry meat employing polymerase chain reaction. M.V.Sc. Thesis submitted to Maharashtra Animal and Fishery Sciences University, Nagpur.
International Standards–ISO (1996). Microbiology of food and animal feeding stuffs– horizontal method for the detection and enumeration of Listeria monocytogenes.1st Edn 11290–1:1 – 15.
Jallewar PK, Kalorey DR, Kurkure NV, Pande VV, Barbuddhe SB (2007). Genotypic characterization of Listeria spp. isolated from fresh water fish. Intl. J. Food Microbiol. 114: 120 – 123.
http://dx.doi.org/10.1016/j.ijfoodmicro.2006.09.034
PMid:17182144
Jamali H, Radmehr B (2013). Frequency, virulence genes and antimicrobial resistance of Listeria spp. isolated from bovine clinical mastitis. The Vet. J. 198(2): 541 – 542.
http://dx.doi.org/10.1016/j.tvjl.2013.06.012
PMid:23880504
Kaur S, Malik SVS, Bhilegaonkar KN, Vaidya VM, Barbuddhe SB (2010). Use of phospholipase–C assay, in vivo pathogenicity assays and PCR in assessing the virulence of Listeria spp. Vet. J. 184: 366 – 370.
http://dx.doi.org/10.1016/j.tvjl.2009.03.032
PMid:19409824
Khan JA, Rathore RS, Khan S, Ahmad I (2013) In vitro detection of pathogenic Listeria monocytogenes from food sources by conventional, molecular and cell culture method. Braz. J. Microbiol. 44(3): 751 – 758.
http://dx.doi.org/10.1590/S1517-83822013000300013
PMid:24516442 PMCid:PMC3910184
Kornspan JD, Rottem S, Nir–Paz R (2014). Cardiolipidsynthetase involved in antagonistic interaction (reverse CAMP phenomenon) of Mycoplasma Species with Staphylococcus aureus beta haemolysis. J. Clin. Microbiol. 52(5): 1622 – 8.
http://dx.doi.org/10.1128/JCM.00037-14
PMid:24599982
Lacier A, de Centorbi ONP (2002). Listeria species in seafoods: isolation and characterization of Listeria spp. from sea food in San Luis, Argentina Food Micrbiol. 19: 645 – 651.
Lammeli UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680 – 685.
http://dx.doi.org/10.1038/227680a0
Liu D (2004) Listeria monocytogenes: comparative interpretation of mouse virulence assay. FEMS Microbiol. Lett. 233: 159 – 64.
http://dx.doi.org/10.1016/j.femsle.2004.02.005
PMid:15043883
Liu L, O'Conner P, Cotter PD, Hill C, Ross RP (2008). Controlling Listeria monocytogenes in cottage cheese through heterologous production of enterocin A by Lactococcus lactis. J. Appl. Microbiol. 104: 1059 – 1066.
http://dx.doi.org/10.1111/j.1365-2672.2007.03640.x
PMid:18005345
Lotfollahi L, Nowrouzi J, Irajian G, Masjedian F, Kazemi B, Eslamian L, Falahat A, Ramez M (2011). Prevalence and antimicrobial resistance profiles of Listeria monocytogenes in spontaneous abortions in humans. Afr. J. Microbiol. Res. 5(14): 1990 – 1993.
Lowry OH, Resoenbrough NS, Farr AL, Randall RJ (1951). Protein measurement with folin–phenol reagent. J. Biol. Chem. 123: 265 – 275.
Mahmoodi MM (2010). Occurrence of Listeria monocytogenes in raw milk and dairy products in Noorabad, Iran. J. Anim. Vet. Adv. 9: 16 – 9.
http://dx.doi.org/10.3923/javaa.2010.16.19
Marquis H, Doshi V, Portnoy DA (1995). The broad–range phospho– lipase C and a metalloprotease mediate listeriolysin O–indepen– dent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect. Immun. 63: 4531 – 4.
PMid:7591098 PMCid:PMC173647
McKellar RC (1994). Use of the CAMP test for identification of Listeria monocytogenes. Appl. Environ. Microbiol. 60: 4219 – 4225.
PMid:7811061 PMCid:PMC201972
Molla B, Yilma R, Alemayehu D (2004). Listeria monocytogenes and other Listeria species in retail meat and milk products in Addis Ababa, Ethiopia. Ethiopian J. Hlth. Dev. 18: 208 – 212.
Momtaz H, Yadollahi S (2013). Molecular characterization of Listeria monocytogenes isolated from fresh seafood samples in Iran. Diag. Pathol. 8: 149.
http://dx.doi.org/10.1186/1746-1596-8-149
PMid:24033984 PMCid:PMC3852293
Moreno LZ, Paixao R, de Gobbi DDS, Raimundo DC, Ferreira TP, Moreno AM, Hofer E, dos Reis CMF, Matte GR, Matte, MH (2014). Phenotypic and genotypic characterization of atypical Listeria monocytogenes and Listeria innocua isolated from swine slaughterhouses and meat markets. J Infect Dev Ctries. 8(4): 416 – 423.
PMid:24727506
Morvan A, Moubareck C, Leclercq A, Herve–Bazin M, Bremont S, Lecuit M, Courvalin P, Le Monnier A (2010). Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob. Agents Chemother. 54(6): 2728 – 2731.
http://dx.doi.org/10.1128/AAC.01557-09
PMid:20385859 PMCid:PMC2876386
Nayak JB, Brahmbhatt MN, Savalia CV, Bhong CD, Roy A, Kalyani IH, Parmar BC (2010). Detection and characterization of Listeria species from buffalo meat. Buff. Bull. 29: 83 – 94.
Notermans SHW, Dufrenne J, Leimeister–Wachter M, Domann E, Chakraborty T (1991). Phosphatidylinositol specific phospholipase C activity as a marker to distinguish between pathogenic and nonpathogenic Listeria species. Appl. Environ. Microbiol. 57: 2666 – 2670.
PMid:1662937 PMCid:PMC183637
Nunes ZG, Hofer E (1994). Evaluation of phenotypic markers associated with pathogenicity in the genus Listeria. Rev. Inst. Med. Trop. Sao Paulo. 36(4): 293 – 299.
http://dx.doi.org/10.1590/S0036-46651994000400001
Osman KM, Samira A, Orabia A, Zolnikovb TR (2014). Confirmed low prevalence of Listeria mastitis in she–camel milk delivers a safe, alternative milk for human consumption. Acta Tropica 130: 1 – 6.
http://dx.doi.org/10.1016/j.actatropica.2013.10.001
PMid:24161878
Park SH, Chang PS, Ryu S, Kang DH (2014). Development of a novel selective and differential medium for the isolation of Listeria monocytogenes. Appl. Environ. Microbiol. 80(3): 1020 – 1025.
http://dx.doi.org/10.1128/AEM.02840-13
PMid:24271177 PMCid:PMC3911231
Parrisius J, Bhakadi S, Roth M, Tranum–Jensen J, Goeber W, Seeliger HPR (1986). Production of Listeriolysin by α–Haemolytic Strains of L. monocytogenes. Infect. Immu. 51(1): 314 – 319.
PMid:3079734 PMCid:PMC261104
Paziak–Domanska B, Boguslawska E, Wieckowska–Szakiel M, Kotlowski R, Rozalska B, Chmiela M, Kur J, Dabrowski W, Rudnicka W (1999). Evaluation of the API test, Phosphatidylinositol–specific phospholipase C activity and PCR method in identification of Listeria monocytogenes in meat foods. FEMS Microbiol. Lett. 171: 209 – 214.
http://dx.doi.org/10.1111/j.1574-6968.1999.tb13434.x
PMid:10077846
Prazak MA, Murano EA, Mercado I, Acuff GR (2002). Antimicrobial resistance of Listeria monocytogenes isolated from cabbage farms and packing sheds in Texas. J. Food. Prot. 65(11): 1796 – 1799.
PMid:12430706
Rahimi E, Ameri M, Momtaz H (2010). Prevalance and antimicrobial resistance of Listeria species isolated from milk and dairy products in Iran. Food Res. Intl. 11: 1448 – 1452.
Raorane A, Doijad S, Katkar S, Pathak A, Poharkar K, Dubal Z, Barbuddhe S. (2014). Prevalence of Listeria spp. in Animals and Associated Environment. Adv. Anim. Vet. Sci. 2(2): 81–85.
http://dx.doi.org/10.14737/journal.aavs/2014/2.2.81.85
Rawool DB, Malik SVS, Barbuddhe SB, Shakuntala I, Aurora R (2007). A multiplex PCR for detection of virulence associated genes in Listeria monocytogenes. Int. J. Food. Saf. 9: 56 – 62.
Rodas–Suarez OR, Flores–Pedroche JF, Betancourt–Rule JM, Quinones– Ramirez EI, Vazquez–Salinas C (2006). Occurrence and Antibiotic Sensitivity of Listeria monocytogenes Strains Isolated from Oysters, Fish and Estuarine Water. Appl. Envir. Microbiol. 72(11): 7410 – 7412.
http://dx.doi.org/10.1128/AEM.00956-06
PMid:16980425 PMCid:PMC1636160
Rosenberg IM (1996). Transfer and detection of Proteins on membranes supports in: Protein analysis and purification. Benchtop Techniques Birkhäuser Boston. 55 – 98.
Ruiz–Bolivar Z, Neuque–Rico MC, Poutou–Pinales RA, Carrascal– Camacho AK and Mattar S (2011). Antimicrobial Susceptibility of Listeria monocytogenes from food Isolates from different cities in colombia. Foodborne Pathog Dis. 8: 913 – 919.
http://dx.doi.org/10.1089/fpd.2010.0813
PMid:21492027
Savini V, Barbarini D, Polakowska K, Gherardi G, Białecka A, Kasprowicz A, Polilli E, Marrollo R, Di Bonaventura G, Fazii P, D'Antonio D, Międzobrodzki J, Carretto E (2013). Methicillin–resis¬tant Staphylococcus pseudintermedius infec¬tion in a bone marrow transplant recipient. J. Clin. Microbiol. 51: 1636 – 1638.
http://dx.doi.org/10.1128/JCM.03310-12
PMid:23486715 PMCid:PMC3647946
Savini V, Paparella A, Serio A, Marrollo R, Carretto E, Fazii P (2014). Staphylococcus pseudintermedius for CAMP–test. Int. J. Clin. Exp. Pathol. 7(4): 1733 – 1734.
PMid:24817971 PMCid:PMC4014255
Schlech WF (2000). Foodborne listeriosis. Clin. Infect. Dis. 31(3): 770 – 775.
http://dx.doi.org/10.1086/314008
PMid:11017828
Shakuntala I, Malik SVS, Barbuddhe SB, Rawool DB (2006). Isolation of Listeria monocytogenes from buffaloes with reproductive disorders and its confirmation by polymerase chain reaction. Vet. Microbiol. 117: 229 – 234.
http://dx.doi.org/10.1016/j.vetmic.2006.06.018
PMid:16860946
Tabouret M, de Rycke J, Dubray G (1992). Analysis of surface proteins of Listeria in relation to species, serovar and pathogenicity. J. Gen. Microbiol. 138: 743 – 753.
http://dx.doi.org/10.1099/00221287-138-4-743
PMid:1588308
Tasci F, Turutoglu H, Ogutcu H (2010). Investigation of Listeria species in milk and silage produced in Burdur province. Kafkas Univ Vet Fak Derg. 16:93 – 97
Todd ECD, Notermans S (2011). Surveillance of listeriosis and its causative pathogen Listeria monocytogenes. Food Control. 22(9): 1484 – 1490.
http://dx.doi.org/10.1016/j.foodcont.2010.07.021
Vasu RK, Sunil B, Latha C, Vrinda MK, Kumar A (2014). Prevalence of Listeria species in meat processing environments. Int. J. Curr. Microbiol. App. Sci. 3(2): 542 – 546.
Wong HN, Chao WL, Lee SJ (1990). Incidence and characterization of Listeria monocytogenes in foods available in Taiwan. Appl. Env. Microbiol. 56(10): 3101 – 3104.
PMid:2126699 PMCid:PMC184905
Yadav MM, Roy A, Bhanderi B, Joshi C (2010). Pheno–genotypic characterization of Listeria monocytogenes from bovine clinical mastitis. Buff. Bull. 29: 29 – 38.
Yucel N, Citak S, Onder M (2005). Prevalence and antibiotic resistance of Listeria species in meat products in Ankara, Turkey. Food Microbiol. 22: 241 – 245.
http://dx.doi.org/10.1016/j.fm.2004.03.007