Advances in Animal and Veterinary Sciences
Research Article
Advances in Animal and Veterinary Sciences 2 (4): 242 – 247Sequence Analysis of the Hemagglutinin–Neuraminidase Gene of Newcastle Disease Virus from Punjab Pakistan
Muhammad Abbas1, 5, Muhammad Zubair Shabbir2, Syed Abdul Khaliq1, 5, Sajjad Ali3, Muhammad Munir4*
- Quality Control Laboratory, Veterinary Research Institute, Lahore 54000, Pakistan
- University of Veterinary and Animal Sciences Lahore 54600, Pakistan; 3Disease Investigation Section, Veterinary Research Institute, Lahore 54000, Pakistan
- Department of Biomedical Sciences and Veterinary Public Health, Swedish University of Agricultural Sciences, 751 89 Uppsala, Sweden
- Livestock and Dairy Development Department, Punjab, Pakistan
*Corresponding author: drmunir.muhammad@gmail.com
ARTICLE CITATION:
Abbas M, Shabbir MZ, Khaliq SA, Ali S, Munir M (2014). Sequence analysis of the hemagglutinin–neuraminidase gene of Newcastle disease virus from Punjab Pakistan. Adv. Anim. Vet. Sci. 2 (4): 242 – 247.
Received: 2014–04–01, Revised: 2014–04–14, Accepted: 2014–04–15
The electronic version of this article is the complete one and can be found online at
(
http://dx.doi.org/10.14737/journal.aavs/2014/2.4.242.247
)
which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
ABSTRACT
Newcastle disease (ND) is a highly contagious disease that results in huge economic losses to the poultry industry worldwide including Pakistan. Despite the disease remained endemic, the genetic nature of the hemagglutinin–neuraminidase (HN) gene of NDV waited investigations in several Asian countries. This study was designed to gain insights into the genetics of HN gene of NDV isolated from Pakistan. Sequence analysis of the HN gene of NDV, isolated from thirteen outbreaks in broiler breeder (n = 3) and broiler chickens (n = 10), was performed. On the basis of clinical symptoms, mean death time (MDT, ~ 47 hours) and intracerebral pathogenicity index (ICPI, 1.5), the isolates were found to be velogenic. The phylogenetic analysis indicated a close relationship studied isolate to the Swedish (Chicken/Sweden/97) and Russian isolates (Sterna/Astr/2755/2001). A parallel comparison of HN gene of Chicken/Pak/2011 with other representative of known genotypes of NDV revealed 15 consistent nucleotide substitutions. Beside these substitutions, important mutations were observed at the crucial sites in the HN protein. This data provide foundation on the evolution of HN gene, possible role of HN gene in disease occurrence and emergence of novel genotypes of NDV.
INTRODUCTION
Newcastle disease (ND), an economically important and transboundary disease, is caused by avian paramyxovirus (APMV) type–1. The Newcastle disease virus (NDV) is enveloped, negative sense ssRNA, and has non–segmented genome of length 15156 bases (Millar and Emmerson, 1988). Based on pathogenicity, three classes of NDV namely lentogenic (apathogenic), mesogenic (intermediate), and velogenic (highly pathogenic) are recognized (Hanson, 1988). However, based on the hypervariable region of the fusion gene, the NDV can be divided into two main classes, I and II. The class II of NDV is further divided into I–XI genotypes (Zhang et al., 2011). The group VI and VII can further be subdivided into sub–genotypes VIa to VI–h (Biagini et al., 1999; Herczeg et al., 1999) and VIIa to VII–f (Munir et al., 2012). Beside their genetic variations, these are characterized as individual antigenic subtypes (Alexander et al., 1997). Genotype VII has been reported to cause a number of outbreaks in East Asia and Western Europe (Munir et al., 2012a; Munir et al., 2010; Munir et al., 2012b). In Pakistan, variants of genotype VII were reported and are considered the most predominant group of NDV currently causing disease both in commercial and rural poultry. Despite of heavy vaccinations, the disease remains endemic in Pakistan, which has raised questions about the possible emergence of antigenic variants that can escape the protective immunity induced by commercial vaccine strains (Cho et al., 2008; Yu et al., 2001). This scenario becomes even complicated in the presence of novel genotypes that we have recently reported in Pakistan (Munir et al., 2012a). Although the current disease upsurge is not investigated at the molecular level, but it is speculated that role of HN protein of NDV should not be overlooked in escaping protecting against filed strain of NDV (Cho et al., 2007).
The HN protein is multifunctional protein that initiates the recognition of cell surfaces receptor (sialic acid), and enhances fusion (F) protein activity on the cell surface, and finally removal of sialic acids from progeny virus resulting into the prevention of viral self–agglutination (Crennell et al., 2000; Kolakofsky et al., 2005). The emergence of antigenic variants with point mutations in the linear neutralizing epitope (positions 345–353) of the hemagglutinin neuraminidase (HN) protein, especially at position 347 has been reported mainly in Korea and China where class II genotype VII viruses (especially VIId) are prevalent and intensive vaccination of poultry has been implemented (Cho et al., 2008; Hu et al., 2010) (10, 11).
This study was conducted to determine the phylogenetic relationship of Pakistani NDV isolates, based on HN gene, to that of previously characterized NDV strains. Additionally, efforts were made to ascertain the pattern of nucleotide substitutions with previously characterized strains, which might explain the role of HN protein in disease protection or occurrence of the epizootics. This is of particular interest when a novel genetic group of NDV has been reported from backyard poultry at northern districts of Punjab province, Pakistan (Munir et al., 2012a; Munir et al., 2010; Munir et al., 2012b). Added to this, the multifunctional properties of HN protein and potential substitutions can be exploited for the designing vaccines against NDV.
MATERIALS AND METHODS
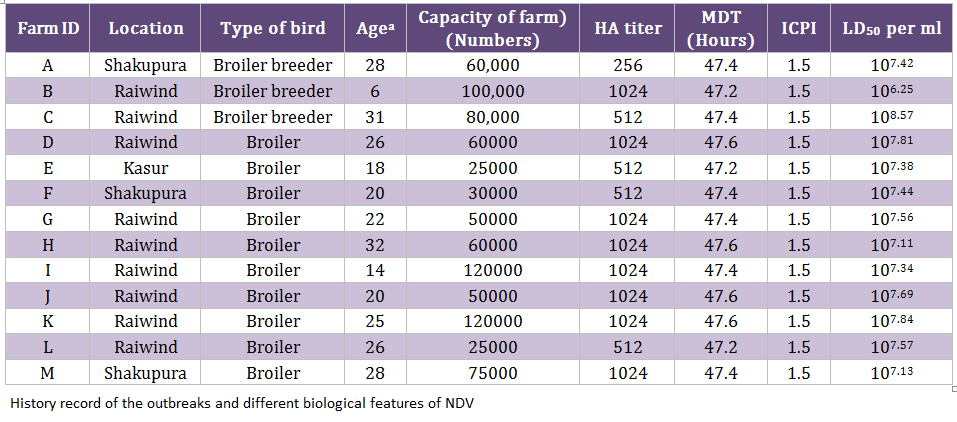
During an epidemic of NDV in 2011/12, 130 samples (trachea, cloaca, and blood) were collected from clinically suspected outbreaks of NDV (n = 13) including broiler breeder (n = 3) and commercial broiler farms (n = 10) (Table 1). A mortality rate of 5–8 percent per day was observed in 6–31 week old breeder broiler flocks, whereas 10–15 percent per day was noticed in 14–32 days old commercial broiler flocks. Collectively, the clinical signs such as dullness, depression, difficult respiration, torticollis and head tremor were noticed. All the infected flocks were vaccinated with commercially available lentogenic strain (LaSota) in the drinking water for commercial broiler birds and killed NDV vaccine for broiler breeder flocks. The tissue homogenates were inoculated in specific pathogen free (SPF) eggs (University Diagnostic Laboratory, UVAS, Lahore, Pakistan) in order to isolate the viruses, as described earlier (Munir et al., 2012a). Mean death time (MDT) and intracerebral pathogenicity indices (ICPI) were calculated as reported earlier (Alexander, 2003; OIE 2004). The egg lethal dose 50 (ELD50) was calculated by Reed and Muench method (Reed and Muench, 1938).
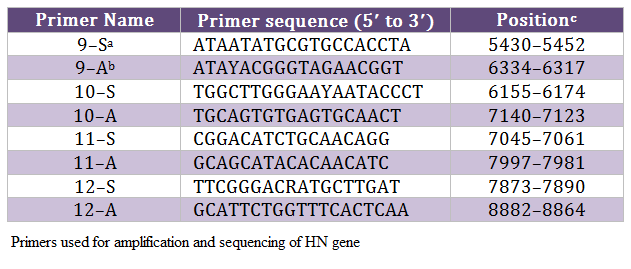
The allantoic fluid from eggs, inoculated with clinical material, was impregnated on FTA QIACard and shipped to Swedish University of Agricultural Sciences (SLU), Uppsala Sweden for further processing. One isolate from each outbreak was selected for analysis due to limited resources. The viral genome was eluted from these cards and run for real–time PCR targeting both matrix (M) and fusion (F) genes, as we described before (Munir et al., 2012a). The positive samples were subjected to conventional PCR in order to amplify the HN gene of NDV, as described before (Munir et al., 2012a), using primers mentioned in Table 2. The amplified PCR products were gel purified with Wizard SV Gel and PCR Clean–Up System (Promega, Co., Madison, WI) according to the manufacturer’s instructions and directly sequenced using ABI PRISM BigDye Terminator version 3.1 (Applied Biosystems, Foster City, CA), as recommended by the manufacturer. Sequencing reactions were run on a 3100 DNA analyzer (Applied Biosystems) both in forward and backward directions. These sequences were assembled and edited in DNAstar Laser gene v8. The consensus sequence was used for the phylogenetic analysis in MEGA5 as we described earlier (Munir et al., 2012a). The consensus sequence of 13 isolates, which show three nucleotides differences, was labeled as Chicken/Pak/2011 and compared with representative strains of each genotype. The final alignment was created in BioEdit. The nucleotide data was submitted to the GenBank under accession number JX436339.
RESULTS
The MDT for all the isolates ranged from 47.2 hours to 47.6 hours whereas the ICPI was observed to be 1.5 (Table 1). Both these values indicated that the isolated viruses were velogenic (3), as was indicated by the clinical signs and mortality. The ELD50 ranged from 106.25 to 108.57/1 mL. The phylogenetic analysis was performed on the basis of nucleotide sequence of the complete open reading frame of HN gene of Chicken/Pak/2011 and 54 NDV strains retrieved from GenBank, which represent all the genotypes of class II. The Phylogenetic tree indicated that representative isolate (Chicken/Pak/2011) clustered with
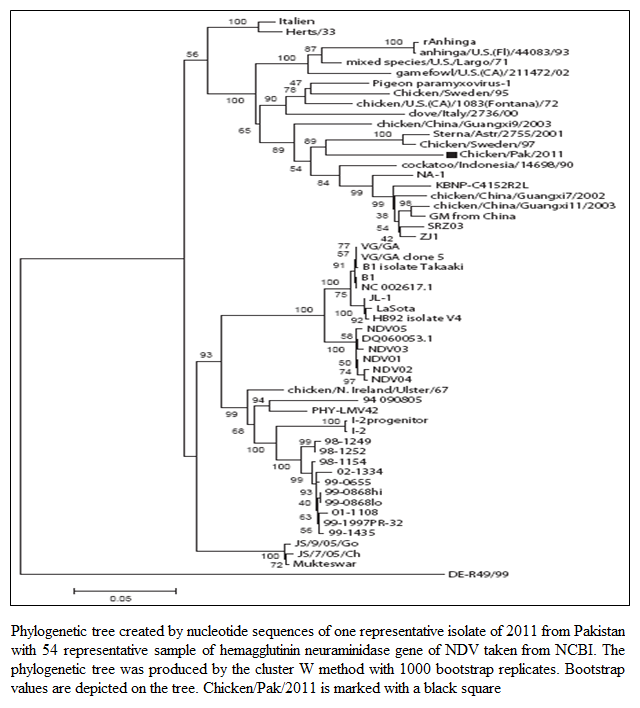
Figure 1: Phylogenetic tree created by nucleotide sequences of one representative isolate of 2011 from Pakistan with 54 representative sample of hemagglutinin neuraminidase gene of NDV taken from NCBI. The phylogenetic tree was produced by the cluster W method with 1000 bootstrap replicates. Bootstrap values are depicted on the tree. Chicken/Pak/2011 is marked with a black square
other strains of NDV in genotype VII within class II (Figure 1). Since all the isolates clustered at the same position, only one isolate was included in the tree. This isolate was found to be closely relate to Swedish (Chicken/Sweden/97, accession number GU585905), and Russian (Sterna/Astr/2755/2001, accession number AY865652) isolates. Both closely related strains were grouped under genotype VII. The Sterna/Astr/2755/2001, was isolated from a Little Tern (Sterna albifrons Pallas), whereas Chicken/Sweden/97 was isolated from an outbreak of ND in commercial poultry form in Sweden (Linde et al., 2010). The isolate, Chicken/Pak/2011, was aligned distinctly, both genetically and phylogenetically, from LaSota and Muktaswar strains, which are currently in use as vaccine strain in Pakistan (Figure 1).
The evolution is a continuous and complex process especially in RNA viruses. Advancement in sequence analysis helps us to understand this process at nucleotide level. Genetic variations during the replication process can be analyzed by sequencing. Mutation can lead to the generation of new genetic groups as has been reported recently from Pakistan (Munir et al., 2012a) and Nigeria (Solomon et al., 2012). To understand such kind of phenomenon, the HN gene was completely sequenced and analyzed. Comparison with prototype strain of NDV that represent each genotype revealed at least 15 nucleotide substitutions for Chicken/Pak/2011. Single nucleotide polymorphism was observed which is given as T6 → C, A33 → G, G79 → A, T108 → C, C140 → G, A145 → G, A169 → G, G319 → A, G333 → T, A405 → G, G450 → A, G507 → A, G530 → A, A590 → G, and C624 → T (Figure 2).
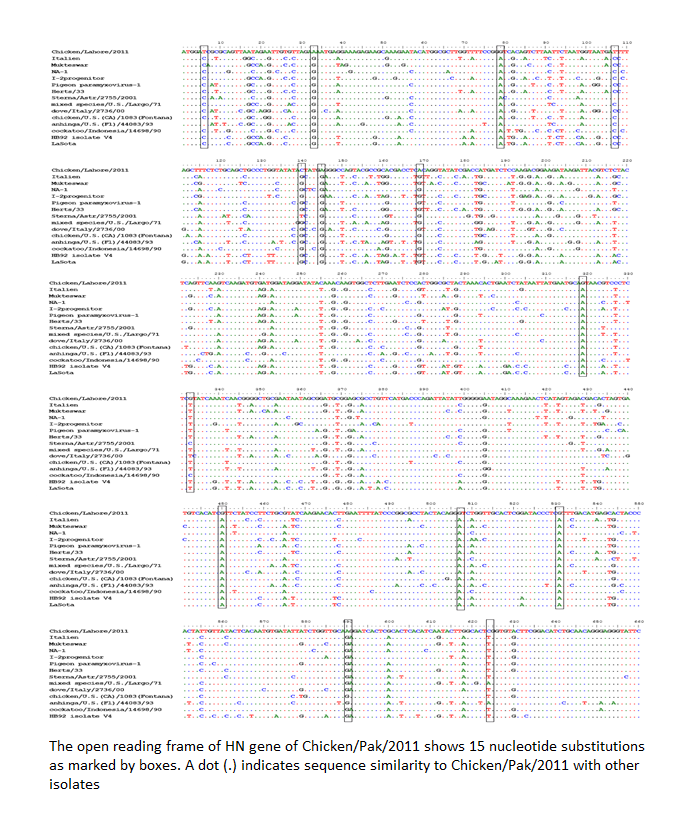
Figure 2: The open reading frame of HN gene of Chicken/Pak/2011 shows 15 nucleotide substitutions as marked by boxes. A dot (.) indicates sequence similarity to Chicken/Pak/2011 with other isolates
Having the crucial role of HN protein in receptor specificity and protective immunity in mind, the analysis of deduced amino acid sequence of HN protein was performed. It has been reported that at least 14 residues are characterized as potential sites for the receptor recognition. These residues are located at 174, 175, 198, 236, 258, 299, 317, 401, 416, 498, 516, 526 and 547 sites. The Chicken/Pak/2011 showed a substitution at 526 (Y526Q) site (Figure 3). Interestingly, Khattar et al., 2009 reported that the NDV strain Beaudette C if mutated at position 526 (Y526Q) reduced the neuraminidase receptor binding and fusion activities of NDV and thus lead to attenuation of viral virulence in eggs and young birds. It has been observed that there are seven antigenic sites within HN protein that are involved in the formation of a continuum in the three dimensional conformation of HN molecules. Out of these sites, substitutions have been observed at I514V, D569V, N263K, E347D, and R353Q sites when compared to LaSota vaccinal
Figure 3: : The crystal structure of HN protein of NDV (PDB ID number 1E8U). The important residues responsible for receptor binding are made sphered. The Y526Q substitution is marked black. The visualization and annotation were carried out using MacPyMole (version 1.3)
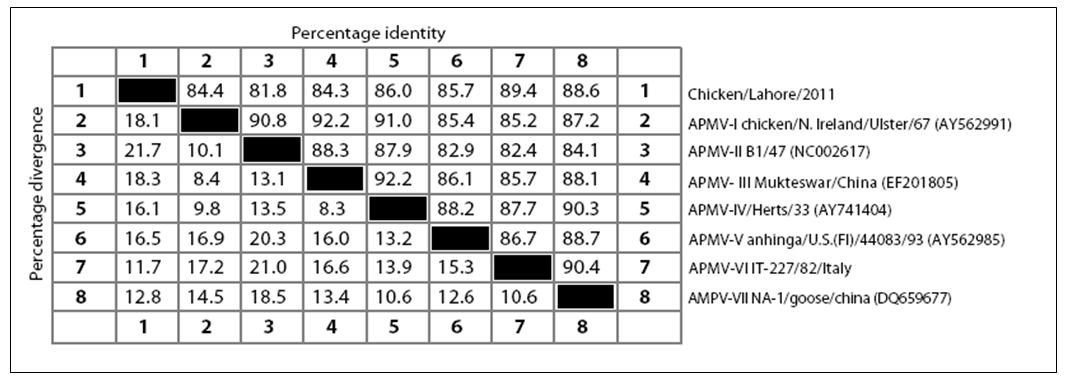
strain. There have been five conserved potential glycosylation sites identified at position 119 (NNSG), 341 (NNTC), 433 (NKTA), 481 (NHTL) and 538 (NKVY) whereas one site at position 508 was absent in Chicken/Pak/2011 which is considered non–conserved among paramyxoviruses (Crennell et al., 2000). The isolated Chicken/Pak/2011 showed a great percentage identity and percentage divergence with other APMV genotypes (I–VII). The Chicken/Pak/2011 showed percentage identity in ascending order with APMV–II, APMV–III, APMV–I, APMV–V, APMV–IV, AMPV–VII and highest percent identity with APMV–VI. Chicken/Pak/2011 showed percentage divergence with other members of APMV in descending order with APMV–VI, AMPV–VII, APMV–IV, APMV–V, APMV–I, APMV– III, and least divergence with APMV–II. There have been three lengths reported for the HN protein of NDV, which depends upon genotype. The lengths of HN protein in genotype I is 616 aa, in genotype II is 577 aa, whereas the HN length in genotypes IV, V and VII are 571 aa, irrespective of their pathogenicity. As expected, being a genotype VII, Chicken/Pak/2011 had a HN length of 571 aa.
DISCUSSION
This study was conducted to investigate the genetic variations in the HN protein of NDV collected from different outbreaks in Pakistan to raise concerns and interest for future investigation in understanding the reasons for recent emergence of the disease. The HN protein, as surface glycoprotein, not only helps in viral attachment to the cell surface receptors but also prevents self–agglutination of the progeny viruses. Therefore, the role of HN protein in protective immunity and escape in vaccine immunity for field virus infections is increasing (Millar and Emmerson, 1988). In line with this Chicken/Pak/2011 isolated from 13 different outbreaks shown same substitution of 15 nucleotides, with a difference of 3 silent nucleotides that don’t affect the nature of putative amino acid. These results dictate that these substitutions are not spontaneous but, being consistent, carry biological meaning and may play crucial role in the occurrence of ND outbreaks. However, such speculations need to be investigated using reverse genetic systems for NDV, which is currently under investigation.
Recently, 23 specific nucleotide substitutions have been reported in the HN gene of NDV from Iran and migratory birds were considered to be important source of transmission especially Sterna albifrons from Russia to Iran (Majid et al., 2012). The virus can be transmitted from Europe to Asia through different species of migratory birds including waterfowls, cranes, teals, pintail, mallard and gadwall (Sheikh et al., 2006). There is a great possibility that the same birds may carry NDV to Pakistan from neighboring country, Iran. In a recent report the role of migratory birds in disease spread by Indus flyway has been discussed (Abbas et al., 2011). Previously genotypes VI and VII have been reported from Karachi, Pakistan, which were closely related to India and China (Khan et al., 2010). In this study, the sequence of HN gene was not determined which left several questions in the evolution of this gene unanswered. Since, the NDV has also been isolated from wild birds of Pakistan including peacock, white partridge and pheasants (M. Munir, unpublished data), the role of migratory and wild birds can be considered as potential source of disease transmission to the commercial poultry.
In Pakistan lentogenic strains of NDV has been used widely for vaccination purposes. In backyard poultry (BYP) Muktaswar strain is being used in the form of live attenuated vaccine for thirty years. During year 2010 and 2011, huge outbreaks of NDV have been reported which facilitated this work to evaluate the evolution of HN gene of NDV. In the current work, isolate Chicken/PAK/2011 showed 15 nucleotide substitutions while comparing with other available HN sequences in GenBank including LaSota (lentogenic) and Muktaswar (mesogenic) strains. The vaccine failure can be correlated with substantial changes at nucleotide level, which results into difference in the amino acids. The results here demonstrated that prevailing field viruses have consistent mutations and substitution at crucial sites. Recently it has been reported from Pakistan that NDV velogenic strain subtype VII–f with substantial mutations in F and HN gene, was producing disease outbreaks in commercial poultry (Munir et al., 2012b). On the other hand, the same NDV velogenic strain subtype VII–f was not producing clinical disease in BYP (Munir et al., 2012c). These findings reflect that the BYP might be the transmission host of NDV to the commercial poultry. The involvement of BYP may also be possible in virus mutation, during virus replication due to presence of high genetic and immune pressure on these viruses. Therefore, these findings are important to evaluate NDV evolution. These substantial changes may lead to alteration in the receptor recognition ability of the virus, resulted into huge out breaks of NDV.
Phylogenetic analysis indicated that the Chicken/Pak/2011 have close relationship with Chicken/Sweden/97 and Sterna/Astr/2755/2001. On the basis of these evidences it can be concluded that the Chicken/Pak/2011 has been originated from the NDV prevalent in Europe particularly from the Sweden Chicken/Sweden/97 and the Russia Sterna/Astr/2755/200. This phylogenetic analysis gives strong evidences of NDV transmission via migratory birds as previously reported (Linde et al., 2010). The Sterna albifrons has also been reported as transmitter of NDV in Russia and Iran (Majid et al., 2012). Furthermore, the prevailing strain of NDV had shown huge phylogenetic gaps with LaSota and Maktaswar. It is reported that high genetic differences and phylogenic gaps among the vaccine strains and field strains enhance evolution (Miller et al., 2007). The evolution is a complex process and its rate is also increasing every year. Therefore, the role of divergent vaccine strains in the emergence of novel genotypes can’t be ignored.
In conclusions, analysis of HN gene from different outbreaks of NDV in commercial poultry farms revealed a consistent pattern of nucleotide, and crucial amino acid substitutions. This data provide foundation on the evolution of HN gene and possible role of HN protein in disease occurrence. Further investigations are required to pinpoint the exact role of these substitutions in the pathogenicity and immunogenicity of the NDV, possibly using reverse genetic systems.
ACKNOWLEDGEMENTS
We would like to acknowledge Director Veterinary Research Institute Lahore, Cantt for help in sample collection.
REFERENCES
Abbas T, Wilking H, Staubach C, Ziller M, Conraths FJ (2011). Priority areas for surveillance and prevention of avian influenzaduring the water–bird migration season in Pakistan. Geospatial Health. 6(1):107–16.
PMid:22109868
Alexander DJ (2003) Newcastle disease virus, other avian paramyxoviruses, and pneumovirus infections. In: Disease of poultry. 11th ed. YM Saif HB, JR Glisson, AM Fadly, LR McDougald, DE Swayne, editor: Iowa State University Press Ames, IA.
Alexander DJ, Manvell RJ, Lowings JP, Frost KM, Collins MS, Russell PH (1997). Antigenic diversity and similarities detected in avian paramyxovirus type 1 (Newcastle disease virus) isolates using monoclonal antibodies. Avian Pathol. 26(2):399–418.
http://dx.doi.org/10.1080/03079459708419222
PMid:18483916
Biagini P, Gallian P, Attoui H, Cantaloube JF, de Micco P, de Lamballerie X (1999). Determination and phylogenetic analysis of partial sequences from TT virus isolates. J. Gen. Virol. 80 (Pt 2):419–24.
PMid:10073702
Cho SH, Kim SJ, Kwon HJ (2007). Genomic sequence of an antigenic variant Newcastle disease virus isolated in Korea. Virus Genes. 35(2):293–302.
http://dx.doi.org/10.1007/s11262-007-0078-z
PMid:17318427
Cho SH, Kwon HJ, Kim TE, Kim JH, Yoo HS, Kim SJ (2008). Variation of a Newcastle disease virus hemagglutinin–neuraminidase linear epitope. J. Clin Microbiol. 46(4):1541–4.
http://dx.doi.org/10.1128/JCM.00187-08
PMid:18272715 PMCid:PMC2292945
Crennell S, Takimoto T, Portner A, Taylor G (2000). Crystal structure of the multifunctional paramyxovirus hemagglutinin–neuraminidase. Nat. Struct. Biol. 7(11):1068–74.
http://dx.doi.org/10.1038/81002
PMid:11062565
Hanson RP (1988). Heterogeneity within strains of Newcastle disease virus: key to survival. Alexander DJ, editor. Boston: Kluwer Academic Publishers.
Herczeg J, Wehmann E, Bragg RR, Travassos Dias PM, Hadjiev G, Werner O (1999). Two novel genetic groups (VIIb and VIII) responsible for recent Newcastle disease outbreaks in Southern Africa, one (VIIb) of which reached Southern Europe. Arch Virol. 144(11): 2087–99.
http://dx.doi.org/10.1007/s007050050624
PMid:10603164
Hu S, Wang T, Liu Y, Meng C, Wang X, Wu Y (2010). Identification of a variable epitope on the Newcastle disease virus hemagglutinin–neuraminidase protein. Vet. Microbiol. 140(1–2):92–7.
http://dx.doi.org/10.1016/j.vetmic.2009.07.029
PMid:19729254
Khan TA, Rue CA, Rehmani SF, Ahmed A, Wasilenko JL, Miller PJ (2010). Phylogenetic and biological characterization of Newcastle disease virus isolates from Pakistan. J. Clin. Microbiol. 48(5):1892–4.
http://dx.doi.org/10.1128/JCM.00148-10
PMid:20237105 PMCid:PMC2863937
Khattar SK, Yan Y, Panda A, Collins PL, Samal SK (2009). A Y526Q mutation in the Newcastle disease virus HN protein reduces its functional activities and attenuates virus replication and pathogenicity. J Virol. 83(15):7779–82.
http://dx.doi.org/10.1128/JVI.00536-09
PMid:19474107 PMCid:PMC2708642
Kolakofsky D, Roux L, Garcin D, Ruigrok RW (2005). Paramyxovirus mRNA editing, the "rule of six" and error catastrophe: a hypothesis. J Gen Virol. 86(Pt7):1869–77.
http://dx.doi.org/10.1099/vir.0.80986-0
PMid:15958664
Linde AM, Munir M, Zohari S, Stahl K, Baule C, Renstrom L (2010). complete genome characterisation of a Newcastle disease virus isolated during an outbreak in Sweden in 1997. Virus Genes. 41(2):165–73.
http://dx.doi.org/10.1007/s11262-010-0498-z
PMid:20640497
Majid E, Mehdi PA, Saber JN, Khadije H (2012). Identification of 23 specific nucleotide patterns in the HN gene of Newcastle disease viruses isolated from Iran. Turk J. Biol. 36:135–42.
Millar NS, Emmerson PT (1988). Molecular cloning and nucleotide sequence of Newcastle disease virus. Alexander DJ, editor. Boston: Kluwer Academic Publishers.
Miller PJ, King DJ, Afonso CL, Suarez DL (2007). Antigenic differences among Newcastle disease virus strains of different genotypes used in vaccine formulation affect viral shedding after a virulent challenge. Vaccine; 25(41):7238–46.
http://dx.doi.org/10.1016/j.vaccine.2007.07.017
PMid:17719150
Munir M, Abbas M, Khan MT, Zohari S, Berg M (2012c). Genomic and biological characterization of a velogenic Newcastle disease virus isolated from a healthy backyard poultry flock in 2010. Virol. J. 9:46.
http://dx.doi.org/10.1186/1743-422X-9-46
PMid:22340092 PMCid:PMC3295720
Munir M, Cortey M, Abbas M, Qureshi ZU, Afzal F, Shabbir MZ (2012a). Biological characterization and phylogenetic analysis of a novel genetic group of Newcastle disease virus isolated from outbreaks in commercial poultry and from backyard poultry flocks in Pakistan. Infect Genet Evol. 12(5):1010–9.
http://dx.doi.org/10.1016/j.meegid.2012.02.015
PMid:22418457
Munir M, Zohari S, Abbas M, Berg M (2012b). Sequencing and analysis of the complete genome of Newcastle disease virus isolated from a commercial poultry farm in 2010. Arch. Virol. 157(4):765–8.
http://dx.doi.org/10.1007/s00705-011-1220-8
PMid:22218968
OIE (2004). Newcastle disease.» Chapter 2.1.15. OIE Manual of standards for diagnostic tests and vaccines, in manual of diagnostic tests and vaccines for terrestrial animals: mammals, birds and bees. Office International des Epizooties, Paris. 270–82.
Reed LJ, Muench LH (1938). A simple method of estimating fifty percent endpoints. American J Hygiene. (27):493–7.
Sheikh KM, Kashif N (2006). Strategic role of Pakistan wetland resources: prospects for an effective migratory waterbird conservation network. Boere GC, Galbraith CA, Stroud DA, editors. Edinburgh, UK: The Stationary Office.
Solomon P, Abolnik C, Joannis TM, Bisschop S (2012). Virulent Newcastle disease virus in Nigeria: identification of a new clade of sub–lineage 5f from livebird markets. Virus Genes. 44(1):98–103.
http://dx.doi.org/10.1007/s11262-011-0678-5
PMid:21960434
Yu L, Wang Z, Jiang Y, Chang L, Kwang J (2001). Characterization of newly emerging Newcastle disease virus isolates from the People's Republic of China and Taiwan. J. Clin. Microbiol. 39(10):3512–9.
http://dx.doi.org/10.1128/JCM.39.10.3512-3519.2001
PMid:11574565 PMCid:PMC88381
Zhang S, Wang X, Zhao C, Liu D, Hu Y, Zhao J (2011). Phylogenetic and pathotypical analysis of two virulent Newcastle disease viruses isolated from domestic ducks in China. PloS One. 6(9):e25000.
http://dx.doi.org/10.1371/journal.pone.0025000
PMid:21949828 PMCid:PMC3176290