Advances in Animal and Veterinary Sciences
Research Article
The Burden and Antibiogram of Methicillin-Resistant Staphylococcus aureus among Companion Animals with Respiratory Illness
Alzahraa R. Attia1*, Khaled A. Abdel-Moein2, Hala M. Zaher2, Ahmed Samir1
1Department of Microbiology, Faculty of Veterinary Medicine, Cairo University, Egypt; 2Department of Zoonoses, Faculty of Veterinary Medicine, Cairo University, Egypt.
Abstract | Methicillin-resistant Staphylococcus aureus (MRSA) is a global pathogen with both veterinary and public health implications. The present study was conducted to investigate the burden of MRSA among diseased companion animals showing respiratory signs. Nasal swabs were collected from 134 companion animals with respiratory illness (48 horses, 41 dogs and 45 cats), all swabs were cultured for MRSA using MRSA CHROMagar medium, whereas isolates were identified as MRSA using conventional methods and molecular detection of mecA gene. Moreover, antimicrobial resistance patterns for all obtained MRSA isolates were determined by the disk diffusion method using the following antibiotics: Cefoxitin, Ceftaroline, Penicillin, Oxacillin, Tetracycline, Doxycycline, Erythromycin, Azithromycin, Clindamycin, Trimethoprim/ sulfamethoxazole, Norfloxacin, Quinupristin/ dalfopristin, Gentamicin and Nitrofurantoin. The prevalence rates of MRSA among horses, dogs and cats were 8.3%, 2.4% and 0% respectively. All the obtained MRSA isolates exhibited multidrug resistance. Two mecA gene sequences obtained in this study (one strain from dog and another from horse) were grouped in the same clade with sequences derived from human patients to underscore the potential public health implications of such strains. In conclusion, the current study highlights the burden of MRSA among diseased companion animals with respiratory illness with special reference to a horse.
Keywords | MRSA, Companion animals, Public health
Received | January 02, 2021; Accepted | June 25, 2021; Published | August 25, 2021
*Correspondence | Alzahraa R. Attia, Department of Microbiology, Faculty of Veterinary Medicine, Cairo University, Egypt; Email: Alzahraa.rabei@gmail.com
Citation | Attia AR, Abdel-Moein KA, Zaher HM, Samir A (2021). The burden and antibiogram of methicillin-resistant Staphylococcus Aureus among companion animals with respiratory illness. Adv. Anim. Vet. Sci. 9(10): 1655-1659.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.10.1655.1659
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Attia et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Companion animals share our life either inside our homes (likewise dogs and cats) or around homes (such as horses). Such animals give us many positive emotions and benefits, but unfortunately, they act as reservoirs for dangerous pathogens which may pass to their owners or human contacts (Abdel-Moein and Samir, 2014). Staphylococcus aureus is a major cause of many infections among humans, ranging from mild to life- threatening conditions likewise sepsis and pneumonia worldwide (Tong et al., 2015). Methicillin-resistant S. aureus (MRSA) has emerged due to the production of an altered penicillin-binding protein (PBP2a) encoded by the mecA gene, such strain was firstly identified in 1961 in the UK (Lakhundi and Zhang, 2018). Afterward, MRSA has been reported as a nosocomial pathogen to stand behind many hospital-acquired infections among humans. Meanwhile, MRSA got its way toward the community and the pathogen waved between people without a history of hospitalization to emerge community-acquired strains of MRSA (Ferreira et al., 2011). On the other hand, several reports have identified MRSA in many infections among animals as well as the pathogen seems to be established among apparently healthy animals and thereby MRSA may have different animal reservoirs to highlight the potential zoonotic risk (Abdel-Moein and Zaher, 2019). The prevalence of MRSA in companion animals was 1.53 % (26/1692) as reported by (Loeffler et al., 2011). Nowadays, MRSA is considered as an emerging pathogen in human and veterinary medicine with climbing public health concerns throughout the world. Moreover, MRSA strains are often multidrug-resistant, making the therapeutic options are limited, thus, MRSA is recognized as one of the most important risks for human and animal health (Morris et. Al., 2017).
The respiratory diseases have a mounting global impact as it is a leading cause of death among humans, especially in middle and low-income countries (FIRS, 2017). Notably, MRSA is incriminated to be a cause of community-acquired pneumonia among humans with severe clinical outcomes (Doudoulakakis et al., 2016; Self et al., 2016).
However, the great attention of MRSA worldwide much remains unknown about the burden of MRSA among the diseased companion animals with respiratory illness. Therefore, the current study was conducted to investigate the burden of MRSA infections among companion animals showing respiratory signs and accordingly tackling the potential zoonotic link to their owners and human contacts.
MATERIALS AND METHODS
Ethical statement
The proposal of this study was approved by the Institutional Animal Care and Use Committee (IACUC) conducted in the Faculty of Veterinary Medicine, Cairo University, Egypt with an ethical approval number: Vet CU16072020171.
Sampling
Nasal swabs were collected from 134 (48 horse, 41 dogs and 45 cats) diseased companion animals showing respiratory signs (coughing, sneezing and nasal discharges). Samples were gathered from different private equine farms, hospitals and pet clinics in Cairo and Giza governorates, Egypt. Sterilized cotton dry swabs were inserted into each nostril for about 2.5cm, rotated for three seconds and repeated for the other nostril (Warren et al., 2004), then inoculated in Amies transport medium (Difco) and transported in an icebox to the laboratory with minimum delay for microbiological processing (Robinson et al., 2012).
Isolation and identification of MRSA
Samples were plated on CHROMagar MRSA medium (CHROMagar, France); which is a chromogenic medium previously evaluated for detection of MRSA with excellent sensitivity and specificity (Diederen et al., 2005). Plates were incubated at 37°C for 48 hours. Suspected colonies were then subcultured on mannitol salt agar medium (Oxoid, UK). Afterward, Gram’s stain films, biochemical tests and coagulase test were carried out according to (Quinn et al., 2011).
Antimicrobial susceptibility testing
All coagulase-positive strains were tested for antimicrobial susceptibility using the disk diffusion method using the following antibiotics: Cefoxitin, Ceftaroline, Penicillin, Oxacillin, Tetracycline, Doxycycline, Erythromycin, Azithromycin, Clindamycin, Trimethoprim/sulfamethoxazole, Norfloxacin, Quinupristin/dalfopristin, Gentamicin and Nitrofurantoin according to the recommendations of Clinical and Laboratory Standards Institute (CLSI, 2018).
Molecular identification of MRSA
All the presumptive cefoxitin-resistant S. aureus strains were subjected to DNA extraction according to (Reischl et al., 1994). Then, S. aureus identification was confirmed by amplification of species specific nuc gene using the following set of primers: (5ʹ TCGCTTGCTATGATTGTGG 3ʹ) and (5ʹ GCCAATGTTCTACCATAGC 3ʹ) (McClure et al., 2017). Each 25μl PCR reaction mixture consisted of 3 μl DNA template, 12.5 μl of PCR master mix (Takara, Japan), 0.5 μl of each primer and final volume was adjusted to 25 μl with 8.5 μl nuclease-free water. The PCR reaction was performed with a thermal profile of 95°C for 3 min, followed by 30 cycles of denaturation, annealing and extension at 95°C for 3o sec, 56°C for 35 sec and 72°C for 1 min respectively then a final extension step at 72°C for 5 min. While confirmation of MRSA strains was achieved by PCR reaction targeting mecA gene using the following set of primers: upstream (5ʹ TGGCTCAGGTACTGCTATCCAC 3ʹ) and downstream (5ʹ AGTTCTGCAGTACCGGATTTGC 3ʹ) (Murakami et al., 1991). Each 25μl PCR reaction mixture consisted of 3 μl DNA template, 12.5 μl of PCR master mix (Takara, Japan), 1 μl of each primer and final volume was adjusted to 25 μl with 7.5 μl nuclease-free water. The PCR reaction was performed with a thermal profile of 94°C for 5 min, followed by 30 cycles of denaturation, annealing and extension at 94°C, 60°C and 72°C for 30 seconds in each step respectively then a final extension step at 72°C for 10 min. The amplicons were analyzed with agarose gel electrophoresis and visualized under a gel documentation system (Biorad, USA) where specific bands were showed at 776 bp (Figure 1).
mecA gene Sequencing
A PCR product of mecA gene of one MRSA isolate from a dog and another from a horse was purified using the QIAquick purification kit (Qiagen, Germany) and sequencing was carried out using Big Dye Terminator V3.1 cycle sequencing kit (Applied Biosystems).
Genbank accession numbers
The two obtained sequences have been deposited in the GenBank database under accession numbers: MN938919 and MN938920.
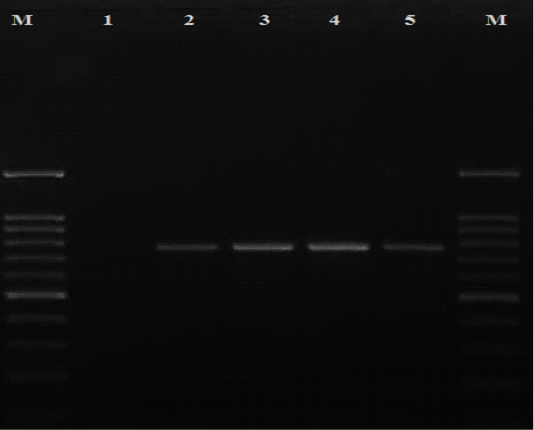
Figure 1: Detection of mecA gene among MRSA isolates recovered in the current study. Lane M: DNA ladder 100 bp; lane 1 negative control (nuclease free water); lanes 2-5 positive isolates for mecA gene with specific bands at 776 bp
Phylogenetic analysis
BLAST analysis was conducted on the obtained sequences to identify similar ones from which some of human and animal origins were retrieved from GenBank. Clustal W Multiple alignments was done to the retrieved sequences by BioEdit software then phylogenetic analysis was carried out using the neighbor-joining method with the MEGA7 software (Figure 2).
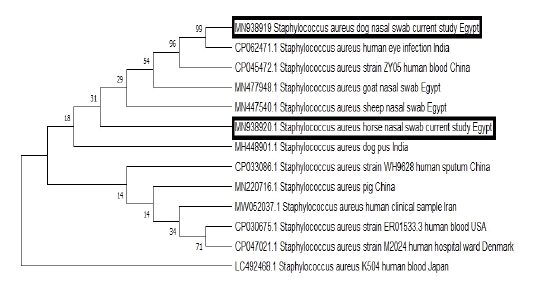
Figure 2: The phylogenetic bootstrap consensus tree demonstrated the evolutionary history of the mecA gene sequences; 2 from the current study and others retrieved from GenBank from human and animal origin. The analysis was conducted using neighbor-joining approach with Mega7 software.
RESULTS and DISCUSSION
The occurrence of MRSA among animals especially pets has been extensively studied (Silva et al., 2020). Several outbreaks in equine veterinary hospitals involved in the transmission of MSRA to horse personnel have been reported (van Duijkeren et al., 2010). Accordingly, the transmission of MRSA between horses, dogs, cats and humans have been highlighted due to the close association of these three animal species and humans (Haenni et al., 2017). The results of the current study revealed that out of 134 examined companion animals with respiratory illness, five animals yielded MRSA giving an overall prevalence rate of 3.7% where the vast majority of isolates were derived from horses (4/48, 8.33%), followed by dogs with a percentage of 2.44 % (1/41) while all the examined cats were MRSA-negative (Table 1). The highest prevalence rate reported among horses (8.3%) was much higher than that obtained by (Hansson et al., 2013) who reported a prevalence rate of 0.1% after examination of horses suffering from respiratory illness.
Table 1: Prevalence of MRSA among diseased companion animals.
| Animal species | No. of examined animals | No. of positive animals | Percentage (%) |
| Horse | 48 | 4 | 8.3 |
| Dog | 41 | 1 | 2.4 |
| Cat | 45 | 0 | 0 |
| Total | 134 | 5 | 3.7 |
On the other hand, the isolation rate of MRSA among the examined dogs was matched with the results obtained by (Abdel-Moein et al., 2012). whereas none of the examined cats yielded positive result to be agreed with those recorded by (Floras et al., 2010; Abdel-Moein et al., 2012) who found that all examined cats were negative for MRSA.
The ascending wave of multidrug-resistant pathogens isolated from companion animals represents a serious burden threatening human health worldwide (Pomba et al., 2017).
Noteworthy, all MRSA isolates recovered in the current study were multi-drug resistant. For horse strains; one strain showed resistance to 78.6% of the applied antibiotics (11/14), other strain was resistant to 71.4% (10/14) while other horse strain was resistant to penicillin, oxacillin, cefoxitin, ceftaroline, doxycycline, erythromycin and nitrofurantoin (50%, 7/14) and the last one was resistant to penicillin, oxacillin, cefoxitin, ceftaroline and tetracycline (35.7%, 5/14). The dog strain was resistant to 71.4% of the tested antibiotics (10/14) as shown in (Table 2).
Surprisingly, all MRSA isolates were found to be resistant to ceftaroline; which is the only beta-lactam antibiotic showing activity against MRSA either in vitro or in vivo. Such antibiotic is considered a drug of choice in some critical and complicated cases of MRSA infections among humans (White et al., 2017; Bhowmick et al., 2019). Strikingly, the results of the current study revealed that two MRSA strains from horses were resistant to Quinupristin/dalfopristin (RP). RP is an antibiotic which that has a high bactericidal activity against drug resistant staphylococci, including MRSA, as such; it may be used as an alternative therapeutic option in certain cases of MRSA infections among human patients with therapy failure (Anwer et al., 1998; Baudoux et al., 2010).
Table 2: The results of antibiotic susceptibility testing of MRSA isolates recovered from companion animals.
| Antibiotic |
Species |
||||
| Dog | Horse | Horse | Horse | Horse | |
| Cefoxitin | R | R | R | R | R |
| Ceftaroline | R | R | R | R | R |
| Penicillin | R | R | R | R | R |
| Oxacillin | R | R | R | R | R |
| Tetracycline | R | R | I | I | R |
| Doxycycline | R | R | R | R | S |
| Erythromycin | R | R | R | R | I |
| Azithromycin | S | R | S | R | S |
| Clindamycin | I | I | S | R | I |
|
Trimethoprim/sulfamethoxazole |
R | S | S | I | S |
| Norfloxacin | R | R | S | R | S |
|
Quinupristin/dalfopristin |
S | R | S | R | S |
| Gentamicin | S | I | S | I | S |
| Nitrofurantoin | R | R | R | S | S |
Resistant (R), susceptible (S), and intermediate (I).
Moreover, one MRSA strain isolated from a dog showed resistance to trimethoprim/sulfamethoxazole which is considered a superior treatment for MRSA pneumonia among humans (Eliakim-Raz et al., 2017).
As mentioned above, the obtained MRSA isolates which were carried by either dog or horses were not only multidrug resistant strains, but also they were resistant to certain antibiotics, which constitute very important therapeutic options for the treatment of critical MRSA infections among human patients. A matter which highlights the public health implication of these strains. Furthermore, the phylogenetic analysis of the obtained sequences from a horse and a dog demonstrated that both sequences were grouped in the same cluster with MRSA strains which were isolated from the blood of human patients and human eye infection from China and India respectively to underline the potential public health importance of such strains and the possible zoonotic link.
CONCLUSIONS AND RECOMMENDATIONS
The current study sheds more light on the burden of MRSA among diseased companion animals with respiratory illness. MRSA should be considered among diseased horses with respiratory illness. Therefore, bacteriological examination of such horses is very important to identify the causative agents and to properly combat them.
NOVELTY STATEMENT
All MRSA strains were resistant to ceftaroline as well as two strains from horse were resistant to Quinupristin/dalfopristin (RP).
AUTHOR’S CONTRIBUTION
Khaled A. Abdel-Moein and Ahmed Samir: Idea, study design, supervising the work, writing manuscript and analysis of data. Alzahraa Rabei: Sample collection, bacteriological examination, molecular techniques and writing manuscript. Hala M. Zaher: Molecular techniques.
Conflict of interest
The authors have declared no conflict of interest.
REFERENCES





