Advances in Animal and Veterinary Sciences
Research Article
Association of OLR1 Polymorphism with Lipid Profile Levels and lipid ratio of Iraqi Awassi Ewes
Mohammed Mahdi Mohammed, Tahreer Mohammed Al-Thuwaini*, Mohammed Baqur Sahib Al-Shuhaib
Department of Animal Production, College of Agriculture, Al-Qasim Green University, Al-Qasim, Babil 51001, Iraq.
Abstract | Oxidized low-density lipoprotein receptor 1 (OLR1) is the major scavenger endothelial receptor that is involved in the binding and degradation of oxidized low-density lipoproteins (Ox-LDL), hence it is responsible in controlling lipid metabolism. Genetic polymorphism in the OLR1 gene is associated with the variation in Ox-LDL of livestock. Therefore, this study aimed to investigate the association of OLR1 polymorphism with the lipid profile levels in Awassi ewes. A total of 200 sexually mature ewes, non-pregnant and non-lactation, aged between 2.5 to 5 years were included in this study. Blood samples were collected from the sheep for extracting genomic DNA, genotyping and the sequencing reaction were subsequently used to confirm the variants in amplified fragments. Blood sera were harvested to determine lipid profile assay. The result referred to the significant differences (P < 0.05) in total cholesterol, HDL-c, LDL-c levels and lipid ratio between the CC genotype compared to the CA genotype. The genotype CC correlated positively with total cholesterol (r= 0.51, P= 0.02), HDL-c (r= 0.63, P= 0.02) and negatively with LDL-c (r= -0.34, P= 0.03), TC/HDL-c (r= -0.43, P= 0.01), LDL-c/HDL-c (r= -0.38, P= 0.04), LDL-c/ TG (r= -0.58, P= 0.02), and TG/HDL-c (r= -0.64, P= 0.02). Sheep with the CC genotype had higher levels of total cholesterol and HDL-c but lower levels of LDL-c and lipid ratio thus are more favored for human consumption. In conclusion, OLR1 gene has a key functions in the regulation of lipid metabolism and affect many economic traits. Based on these outcomes, OLR1 gene represented one of the important candidate genes for fat deposition-related traits in livestock.
Keywords | Lipid profile; Ox-LDL; OLR1; polymorphism; sheep.
Received | May 14, 2021; Accepted | June 12, 2021; Published | July 28, 2021
*Correspondence | Tahreer Mohammed Al-Thuwaini, Department of Animal Production, College of Agriculture, Al-Qasim Green University, Al-Qasim, Babil 51001, Iraq; Email: tahreermohammed@ymail.com
Citation | Mohammed MM, Al-Thuwaini TM, Al-Shuhaib MBS (2021). Association of olr1 polymorphism with lipid profile levels and lipid ratio of iraqi awassi ewes. Adv. Anim. Vet. Sci. 9(9): 1460-1465.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.9.1460.1465
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Al-Thuwaini et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Lipids play several roles in the animal body as hormones, storage, insulation, participate in cell membranes component and determine the milk and meat quality in livestock (Adamu et al., 2008; Hocquette et al., 2010). Although these important roles but any changes in the plasma lipid levels or in the fat content of milk or meat could lead to many clinical disorders (Khaki et al., 2012). Excessive consumption of lamb meat with high-fat content can lead to several chronic diseases (Hatta et al., 2019). So many efforts have been made to reduce the fat content by using modern genetic technologies (Gorlov et al., 2020). One of the most important candidate genes is oxidized low-density lipoprotein receptor 1 (OLR1) which is located on chromosome 3 in sheep and is consisting of 6 exons (Adopted from ncbi.nlm.nih.gov). In cattle, it exists on chromosome 5 and is consisting of 5 exons (Anggraeni, 2019). OLR1 gene encodes oxidized low-density lipoprotein receptors protein (OLR1) belongs to the C-type lectin family that functions to absorbs and degrade low-density lipoproteins (Kowalewska-Łuczak and Czerniawska-Piątkowska, 2018).
OLR1 is a type II membrane protein (50 kDa) that represent the major cell-surface scavenger receptor for oxidized low-density lipoprotein (LDL) on endothelial cells (Murphy et al., 2005; Soltani-Ghombavani et al., 2013). LDL-c is very sensitive to oxidative damage and is converted to harmful oxidized LDL lipid (ox-LDL), which is elevated in the plasma of cardiovascular patients (Hendrikx et al., 2014). The accumulation of oxidized LDL in the coronary arteries before the development of coronary plaques suggests that increased levels of oxidized LDL are a marker for early atherosclerosis (Holvoet et al., 2001). A more recently, oxidized LDL and lipid indexes such as total cholesterol/ high density lipoprotein-cholesterol (TC/HDL-c), low density lipoprotein-cholesterol / high density lipoprotein-cholesterol (LDL-c/HDL-c) and triglycerides/ high density lipoprotein-cholesterol (TG/HDL-c) are considered to be good parameters for predicting cardiovascular risk, as they may provide an integrated set of information based on multiple variables (Guo et al., 2019). The use of these indicator can be more valuable than lipid levels alone and can provide information on risk factors that are difficult to quantify by routine testing, and may better reflect the clinical and metabolic interactions between disorders of lipid metabolism (Baez-Duarte et al., 2017; Hegazy et al., 2020). OLR1 is recognized oxidation of LDL and then triggers the transformation of macrophages during plaque formation and cellular apoptosis (Murphy et al., 2008). Hence, a non-functional OLR1 SNP results in modification of oxidation of LDL and are thought to play a key role in the pro-inflammatory conditions and arteriosclerosis development (Khan et al., 2015; Jamatia et al., 2018). Many researchers reported the polymorphism of the OLR1 gene in livestock and its association with economic traits. Khatib et al. (2006); Komisarek and Dorynek (2009); Vinsky et al. (2013); Fonseca et al. (2015); Kowalewska-Łuczak and Czerniawska-Piątkowska (2018). Anggraeni (2019) reported that the SNP in the coding region or non-coding region of the OLR1 gene is associated with a significant effect on economics traits in cattle. Javed et al. (2013) revealed the association of polymorphism of the OLR1 gene with growth traits and milk fat content in buffalo. Based on the aforementioned studies, no studies related the association of OLR1 gene with the lipid profile levels of Awassi sheep had been undertaken. Therefore, this study aimed to investigate the association of OLR1 polymorphism with lipid profile levels in Awassi ewes.
MATERIALS AND METHODS
Animals and experimental design
This study was approved and conducted at the Al-Qasim Green University’s according to the international recommendations for the care and use of animals with approval number
(Agri, No. 020,7,18), for the period from July 2020 to March 2021 on Awassi ewes. A total of 200 sexually mature ewes, non-pregnant and non-lactation, aged between 2.5 to 5 years were included in this study. Animals were collected randomly from two stations for raising sheep (Babylon and Karbala). Ewes were fed ad libitum on seasonal grass, concentrate food, and fresh water. Blood samples (5 ml) were collected in the morning before feeding from the jugular vein of the sheep using vacutainer tubes with an anticoagulation tube for genetic analysis and in blank clean test tube for performing lipid profile. Blood samples without anticoagulant were centrifuged at 3,000 rpm at room temperature for 15 min to collect the sera. The obtained sera were transferred into Eppendorf tubes and it was kept frozen at –20 °C to estimate lipid profile assay.
Biochemical examination
Lipid profile analyses were performed according to the instructions of RANDOX Laboratories kits. Total cholesterol and triglycerides were analyzed by using the colorimetric enzymatic method and by using the precipitant method for HDL-cholesterol. LDL-cholesterol was calculated using the equation of Friedewald et al. (1972): LDL-Cholesterol = Total Cholesterol – Triglycerides/2.2 – HDL-Cholesterol. Additionally, TC/HDL-c, LDL-c/HDL-c, TG/HDL-c, LDL-c/TG were calculated in this study.
DNA extraction, genotyping and sequencing reaction
Genomic DNA was extracted from the whole blood using a rapid salting-out procedure (Al-Shuhaib, 2017). Briefly, blood was mixed with distilled water to destroy red blood cells. Resuspend the pelleted leukocytes in lysis buffer (10 mM Tris-Cl pH 7.7, 0.4 M NaCl, 2 mM EDTA, 0.5% SDS). The protein is denatured by 6 M NaCl. The supernatant containing DNA was precipitated with absolute ethanol. The precipitation DNA strands were washed with 70% ethanol. DNA recovery via elution buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). The extracted DNA samples were evaluated uses NanodropμLITE spectrophotometer (Biodrop, UK) and used as a template for polymerase chain reaction (PCR). The PCR reaction contained (10 pmol of each primer, 50 ng of genomic DNA, 50 μM dNTPs, 10mM Tris-HCl (pH 9.0), 30mM KCl, 1.5mM MgCl2, and 1 U Top DNA polymerase). The primer designed for the OLR1 gene (exon 3) based on the GenBank ovine sequence (NC_040254.1) and Primer-BLAST. The sequences of the forward and reverse primers were 5 ́- TTCACCGAAATCCAGTGGAGG -3 ́ and 5 ́- CCTCTCGCCCTCATACCTGA -3 ́ respectively. The PCR amplification was initial by denaturation at 94°C for 4 min, 35 cycles of denaturation at 94°C for 30 s, 57°C annealing temperature for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 10 min. The PCR products were run on 2 % agarose gel and visualized under UV light.
Post PCR, genotyping was performed using the polymerase chain reaction-SSCP (PCR-SSCP) technique (Imran et al., 2020). Each PCR amplicon was treated with an equal volume of SSCP denaturing-loading buffer. After denaturation for 7 min and chilled on wet ice for 10 min, the denatured samples were loaded on neutral polyacrylamide gels in 0.5 TBE buffer. Electrophoresis conditions were carried out on a constant 200 mA and 100 V for 4 hrs at room temperature. Gels were stained according to the rapid protocol described by Byun et al. (2009). The nucleic acid polymorphisms in each detected genotype were sequencing in (Macrogen, Geumchen, Korea), then edited and visualized by BioEdit ver, 7.1. (DNASTAR, Madison) and SnapGene Viewer ver. 4.0.4 (http://www.snapgene.com). Moreover, the Ensemble genome browser 96 (https://asia.ensembl.org/index.html) was used to check the novelty of the observed variants of the OLR1 gene.
Statistical analysis
The association of OLR1 genotypes with traits of interest was analyzed through the general linear model (GLM) procedure by SPSS (version 23.0) with the following model:
Y ijk = μ + Gi + Pj + Ak + eijk
where Y ijk = value for studied traits, μ = overall mean, Gi = fixed effect of ith genotypes (i = CC, CA), Pj = fixed effect of jth parity (j = 1, 2, 3, 4), Ak = fixed effect of kth age group (2.5-3.5,>3.5-5), and e ijk = random error associated with Y ijk observation and assumed to be NID (0, σ2e). Means were compared using Tukey-Krammer test. Normality was tested using the Kolmogorov–Smirnov test. The correlation was analyzed using the Pearson correlation coefficient, and significance was set at P < 0.05. Preliminary statistical analysis indicated the effect of factor interaction, season and nutrition did not have a significant effect on studied traits, so they were excluded from the model.
RESULTS AND DISCUSSION
Association analysis of OLR1 genotypes with lipid profile levels of Awassi ewes
Polymerase chain reaction (PCR) experiments were conducted for all 200 samples (114 ewes with singleton births and 86 ewes with twin’s births; Figure 1). Genotyping technique detected two distinct PCR-SSCP banding patterns CC and CA genotypes (Figure 2); sequencing reaction confirmed a novel mutation C246A SNP. Regarding the effects of OLR1 genotypes on lipid profile levels and lipid ratio of Awassi ewes, the result showed a significant differences (P < 0.05) in total cholesterol, HDL-c, LDL-c levels and lipid ratio between CC genotype compared to CA genotype as shown in (Table 1).
Table 1: Relationship of OLR1 genotypes with lipid profile levels and lipid ratio of Awassi ewes.
| Indices |
OLR1 Genotype (LSM ± SE) |
P-value |
|
|
CC (51) |
CA (149) | ||
| Total cholesterol (mg/dl) |
91.42 ± 2.31 a |
81.71 ± 2.53 b |
0.02 |
| Triglyceride (mg/dl) |
36.74 ± 1.56 a |
34.11 ± 1.74 a |
0.43 |
| HDL-c (mg/dl) |
46.34 ± 2.14 a |
31.12 ± 1.82 b |
0.03 |
| LDL-c (mg/dl) |
32.62 ± 0.81 a |
49.34 ± 0.60 b |
0.04 |
| TC/HDL-c | 2.28 ± 0.13 | 2.95 ± 0.11 | 0.001 |
| LDL-c/HDL-c | 0.82 ± 0.06 | 1.74 ± 0.04 | 0.001 |
| LDL-c/ TG | 0.91 ± 0.02 | 1.53 ± 0.05 | 0.001 |
| TG/HDL-c | 0.87 ± 0.07 | 1.12 ± 0.09 |
0.009 |
LSM ± SE, Least square means ± Standard error. Different superscript in the same raw within each classification indicates significant differences (P<0.05). LDL, Low-density lipoprotein, HDL, High-density lipoprotein, TG, Triglycerides.
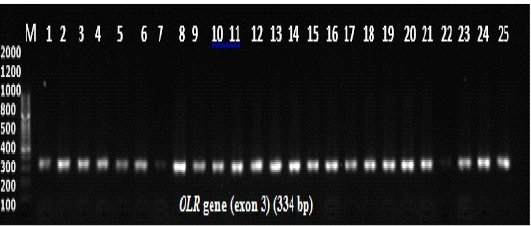
Figure 1: Agarose gel electrophoresis for PCR products amplification of the OLR1 gene. Lane M: 100 bp DNA ladder, lanes 1-12 indicate samples of ewes with single birth, lanes 13-25 indicate samples of ewes with twin birth. Electrophoresis conditions: 1.5% agarose concentration, Electrophoresis time 25 minutes. The buffer used TBE pH 8.3.
The CC genotypes of the OLR1 gene was associated with the highest total cholesterol and HDL-c levels and the

Figure 2: SSCP non-denaturing polyacrylamide gel electrophoresis of ovine OLR1 gene (exon 3) PCR fragments, which appeared two PCR-SSCP banding patterns, CC and AC in Awassi ewes.
lowest LDL-c levels and lipid ratio. This means that this genotype is more controlling on lipid metabolism. It is reported that the oxidized LDL correlates with other lipid variables. The ratio of LDL-c/HDL-c is decreased by an increased LDL oxidation (Shittu et al., 2007). As a core surface scavenger receptor, OLR1 is reported to internalize and degrade Ox-LDL, thereby controlling lipid metabolism and decreased lipid ratio (Shittu et al., 2007; Gui et al., 2019). Jamatia et al. (2018) mentioned that the type of OLR1 SNP was determined the OLR1 gene expression and the role of OLR1 in lipid metabolism, in which the functional OLR1 SNP associated with lower ox-LDL and ultimately affecting animal phenotypes. Similarly, SNP (C223A) in 3’UTR of OLR1 gene in Polish Holstein-Friesian bulls and Holstein Friesian dairy cattle resulted in higher milk fat content and lower ox-LDL to the CC and AC genotypes compared to AA genotypes (Komisarek and Dorynek, 2009; Anggraeni, 2019).
Correlation analysis of OLR1 genotypes with lipid profile of Awassi ewes
The correlation coefficient between OLR1 genotypes with lipid profile levels and lipid ratio of the Awassi ewes are presented in Table 2. The genotype CC were positively correlated with total cholesterol (r= 0.51, P= 0.02), HDL-c (r= 0.63, P= 0.02), and negatively with LDL-c (r= -0.34, P= 0.03), TC/HDL-c (r= -0.43, P= 0.01), LDL-c/HDL-c (r= -0.38, P= 0.04), LDL-c/ TG (r= -0.58, P= 0.02), and TG/HDL-c (r= -0.64, P= 0.02). There was a non-significant (P > 0.05) correlation between the genotype CC and the triglyceride.
The result indicated the presence of a positive and significant correlation (P< 0.05) between CC genotype and lipid profile levels and negatively with LDL-c and lipid ratio. The ratio TG/HDL-c is correlated inversely with the plasma level of small dense LDL particles (da Luz et al., 2008). Higher TG and lower HDL-C, as evidenced by an increased TG/HDL-C ratio, thus potentially contribute to inflammation, a risk factor for cardiovascular disease (Anderson et al., 2021). Moreover, an elevated LDL-C/
Table 2: Correlation between OLR1 genotypes with lipid profile and lipid ratio in Awassi ewes
| Variables |
OLR1 genotype |
|||
| CC | CA | |||
| r |
P-value |
r |
P-value |
|
| Total cholesterol | 0.51 | 0.02 | 0.41 | 0.04 |
|
Triglyceride |
0.19 | 0.21 | 0.28 | 0.45 |
| HDL-c | 0.63 | 0.02 | 0.30 | 0.03 |
| LDL-c | -0.34 | 0.03 | -0.25 | 0.04 |
| TC/HDL-c | -0.43 | 0.01 | -0.42 | 0.03 |
| LDL-c/HDL-c | - 0.38 | 0.04 | 0.48 | 0.05 |
| LDL-c/ TG | -0.58 | 0.02 | -0.66 | 0.04 |
| TG/HDL-c | -0.64 | 0.02 | -0.45 | 0.04 |
HDL-C and TC/HDL-C ratio combined with elevated TG is associated with a high cardiovascular risk (Lemieux et al., 2001). The ratio of LDL-c/HDL-c is decreased by an increased LDL oxidation, which is a better indicator of lipid peroxidation (Shittu et al., 2007). OLR1 is responsible for the internalization and degradation of ox-LDL and decreasing lipid ratio thus ultimately affecting animal traits (Jamatia et al., 2018). This result is consistent with the study of Fonseca et al. (2015) that reported a significant correlation (P ≤ 0.05) between CC genotype in OLR1 gene with growth and carcass traits in Nelore cattle. Similarly, Khatib et al. (2006) and Soltani-Ghombavani et al. (2013) observed a relationship of OLR1 polymorphism with lipid metabolism in Holstein dairy cattle and dairy cows. Although this gene has been proven to significantly influence on economic traits of livestock (Fonseca et al., 2015; Kowalewska-Łuczak and Czerniawska-Piątkowska, 2018; Anggraeni, 2019). However, no studies related to the association of the OLR1 gene with the lipid profile levels and economic traits of Awassi sheep had been undertaken. Hence, genotypic information and novel associations of this study can be more useful in selecting sheep with low levels of LDL and it can be recommended for the assessment of functional traits in future marker-assisted selection programs.
STUDY LIMITATIONS
This study had three main limitations. Undoubtedly, the small size of the studied population is still a limitation of our research. Second, this study focused on Awassi sheep, therefore, the conclusions cannot be extrapolated to different sheep breeds. Third, the absence of data such as economic traits may have a potential influence on outcomes.
CONCLUSIONS
OLR1 gene polymorphism associated key functions in the regulation of lipid metabolism, meat and milk fat content. Based on these outcomes, the OLR1 gene represents one of the important candidate genes for fat deposition-related traits in livestock. OLR1 gene polymorphism could be used in selecting sheep that produce body fat with low levels of LDL and lipid ratio. Sheep with OLR1 gene polymorphism should be selected and future studies relating this gene with high twining rate are required in the future.
ACKNOWLEDGEMENT
The authors are grateful to the staff of two stations for raising sheep (Babylon and Karbala) for their facilities that provided the Awassi ewes population.
CONFLICT OF INTEREST
There is no conflict of interest.
AUTHORS CONTRIBUTION
All authors contributed equally.
REFERENCES





