Advances in Animal and Veterinary Sciences
Research Article
Comparative Diagnostic Performance of Microscopic Examination, Polyclonal Antigen-ELISA, and Polymerase Chain Reaction for the Detection of Trypanosoma evansi in Camels (Camelus dromedarius)
Arafat Sadek1, Khaled A.S. El-Khabaz2*, Sherief M. El-Genedy3, Magdy M. El-Gioushy4
1Department of Animal Medicine (Internal Medicine), Faculty of Veterinary Medicine, Assiut university, Assiut - Egypt; 2Department of Animal Medicine (Infectious Diseases), Faculty of Veterinary Medicine, Assiut University, Assiut - Egypt; 3Field Practitioner, Assiut - Egypt; 4Department of Animal Medicine (Internal Medicine), Faculty of Veterinary Medicine, Aswan University, Aswan-Egypt 37916, Egypt.
Abstract | Camel trypanosomiasis (surra) is a vector-borne parasitic disease caused by Trypanosoma (T.) evansi, affecting the health of camels, and resulting in a number of negative economic implications. The clinical signs of the disease are not characteristic, and as such, the laboratory diagnosis of camel trypanosomiasis is crucial. This study aimed to demonstrate the diagnostic performance of Giemsa-stained blood smears (GSBS), antigen enzyme-linked immunosorbent assay (Ag-ELISA) to polymerase chain reaction (PCR) for the diagnosis of T. evansi infection in camels. Blood samples were collected from 56 clinically suspected and 36 apparently healthy camels. The overall prevalence of T.evansi was 9.8 % with GSBS with sensitivity 16.7% and specificity 100%, 16.3 % with Ag-ELISA with sensitivity 27.8% and specificity100%, and 58.7 %with PCR. The prevalence of T. evansi in the clinically suspected camels was 16.1% with GSBS, 26.8% with Ag-ELISA, and 60.7% with PCR, while in the apparently healthy camels, was 55.6% with PCR and no positive cases were detected with both GSBS and Ag-ELISA. A significant increase in the prevalence identified with GSBS, Ag-ELISA, and PCR was found in clinically suspected camels relative to the apparently healthy ones. A substantial agreement (k=0.715) between GSBS and antigen-ELISA was detected, while the agreement between GSBS and PCR was slight (k =0.142) and fair (k =0.241) between Ag-ELISA and PCR. This study advocates the use of molecular analysis to assess the prevalence of surra in camels, rather than antigen-ELISA. However, GSBS can be used in developing countries as a preliminary screening method.
Keywords | Camel, Trypanosomiasis, Blood smear, PCR, ELISA
Received | March 30, 2021; Accepted | April 10, 2021; Published | June 15, 2021
*Correspondence | Khaled A.S El-Khabaz, Department of Animal Medicine (Infectious Diseases), Faculty of Veterinary Medicine, Assiut University, Assiut - Egypt; Email: khaled.sayed@aun.edu.eg
Citation | Sadek A, El-Khabaz KAS, El-Genedy SM, El-Gioushy MM (2021). Comparative diagnostic performance of microscopic examination, polyclonal antigen-elisa, and polymerase chain reaction for the detection of trypanosoma evansi in camels (camelus dromedarius) Adv. Anim. Vet. Sci. 9(7): 1004-1011.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.7.1004.1011
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 El-Khabaz et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Camels in Arab countries are of concern, as they are an important part of Arab culture and economy. They are well-adapted to the hot and arid desert climate. Camels have a multi-functional role in grain and water transportation, as well as meat and milk production (Samara et al., 2012; Pasha et al., 2013). The world’s camel population is gradually growing in many parts of the world, particularly the Arab countries (Wernery and Kaaden, 2002). Camels were previously regarded as resistant to many infectious diseases, but recently a variety of bacterial, viral, and parasitic diseases have been reported (Kassa et al., 2011).
Surra is an enzootic protozoal disease of camels in Egypt caused by T. evansi (Sobhy et al., 2017). Mechanical transmission of the T.evansi occurs by haematophagus flies such as Tabanus, and Stomoxys (Aregawi et al., 2019). Surra can occur with or without clinical signs (Hassan-Kadle et al., 2019). Most of the reported clinical cases were chronic with anemia and intermittent fever, but there were also documented acute cases that could be fatal in weeks (Desquesnes et al., 2013). The clinical signs of camel trypanosomiasis are not adequate on their own to diagnose the disease, since they are non–specific. Moreover, asymptomatic infection may occur. Therefore, laboratory diagnosis is required for a conclusive diagnosis (Cadioli et al., 2012).
Parasitological methods such as microscopic examination of wet blood or stained blood films are used for laboratory diagnoses of the parasites (Chagas et al., 2020). Despite the low sensitivity of these tests due to intermittent parasitemia, and the low number of parasites in the chronic cases, they are routinely used since they are cheap and fast (Yadvendra et al., 1998). Parasite concentration techniques, such as microhematocrit centrifugation and buffy coat method, are also used to detect motile and live trypanosomes (Nuryady et al., 2019), but these techniques are incapable of identifying trypanosome species (Muieed et al., 2011). Many serological tests are used for screening of camel trypanosomiasis, including an antigen-ELISA test for detection of parasitic antigen (Sengupta et al., 2019) or antibody–ELISA (Sivajothi et al., 2016), indirect fluorescent antibodies (Aquino et al., 2010), and card agglutination tests (Songa and Hamers, 1988) for antibody detection. The serological tests that detect antibodies cannot distinguish between past and recent infections (Tehseen et al., 2015), while those that detect antigens cannot provide adequate sensitivity (Desquesnes, 1996). Many molecular tests, such as conventional and real time PCR have been used to diagnose camel trypanosomiasis through the detection of trypanosomal DNA. These molecular tests are more sensitive than other techniques and have the advantage of being capable of classifying parasites at the subspecies level (Barghash et al., 2014; Elhaig et al., 2013). Early and accurate diagnosis of T.evansi is a prerequisite for disease control as it will improve treatment efficacy, reduce long term complications, and prevent further spread of the disease by undiagnosed cases. Additionally, the limitations of many diagnostic techniques necessitate the use of an accurate diagnostic method. Therefore, the current study aimed at evaluating conventional diagnostic methods in comparison to recent techniques for the diagnosis of camel trypanosomiasis to find a suitable diagnostic method.
Materials and Methods
Animals
This study was conducted on 92 one-humped male camels (Camelus dromedarius) of various ages from different localities in the Aswan governorate. Fifty-six camels had suspected clinical signs of camel trypanosomiasis and 36 appeared to be healthy.
Blood samples
Venous blood samples were collected from all camels enrolled in this study. Five ml of blood was collected in EDTA coated-vacutainer tubes, for parasitological analysis, which was then stored at- 20°C for DNA extraction for PCR. Five ml of blood was drained in the vacutainer tubes to separate serum, blood coagulation that occurred within 20 minutes at room temperature, followed by centrifugation at 3000 rpm for 20 minutes. The supernatants were transferred to the Eppendorf tubes using Pasteur pipettes. Serum samples were maintained at -20 °C until they were used for the detection of Trypanosoma antigen using the antigen-ELISA test.
Clinical examination
All Camels enrolled in this study were subjected to a thorough clinical examination as reported previously by (Higgins, 1983). Body temperature, heart, and respiratory rate, mucous membrane, lymph nodes, and muscles of the thigh were reported.
Parasitological diagnosis using GSBS
Stained blood smears were used for parasitological diagnosis of T. evansi according to (Coles, 1986). On one end of a clean microscope slide, a drop of blood was placed and a smear is drawn out. Air-dried blood smears were fixed in absolute methyl alcohol for 3 minutes and allowed to dry again, then stained for 30–45 minutes by Giemsa stain 10%. Microscopic detection of trypanosomes using an oil immersion lens at100x magnification was conducted.
Polyclonal Antigen-Enzyme-Linked Immunosorbent Assay (Antigen -ELISA)
A commercially available kit of Glory was used to identify the antigens of trypanosomes in camel serum according to the method previously recommended by (Clausen et al., 2003). The test procedure was done according to the instructions of the manufacturer as the following; a fifty μl of positive control and negative control were added to the wells (controls were tested in duplicate). Forty μl of sample diluent and 10 μl of serum were added to the other wells and gentle mixing was conducted. The plate was coated with adhesive strips and incubated at 37 °C for 30 minutes. The adhesive strips have been exposed and the liquid was poured out from the wells. Each well washed with 30 seconds-long diluted washing buffer, and then the solution was drained, this process was repeated 5 times. Fifty μl of Horseradish Peroxidase (HRP)-Conjugate reagent was applied to each, followed by incubation at 37 °C for 30 minutes after covering the plate with adhesive strips. After washing the wells for 5 times as previously reported, 50 μl of Chromogen solution A and 50 μl of Chromogen solution B were added to each well, incubation was carried out for 15 min at 37 ℃ away from light. After adding 50μl of stop solution to each well to stop the reaction, the wells have been read with ELISA reader at 450 nm within 15 min.
DNA extraction and PCR detections of Trypanosoma evansi
Whole blood samples were used for DNA extraction with a commercial kit (Qiagen DNeasy blood and tissue kit) as instructed by the manufacturer. The PCR amplification of 164 bp was conducted following the method defined by (Masiga et al., 1992). A specific primer set of highly repeated sequences of mini-chromosome satellite DNA (TBr1 & TBr2) (TBR1: 5’- GAA TATTAAACAATGCGCAG -3’ and TBR2: 5’-CCATTTATTAGCTTTGTTGC-3’) that commercially prepared was used. PCR amplification was started by the initial denaturation step at 94 °C for 10 min, followed by 40 cycles consisting of denaturation for 30 sec. at 94°C. Annealing at 46 °C for 45 sec. and extension at 72 °C for 45 sec were also performed. Final extension step was at 72 ° C for 10 min. Electrophoresis of the amplification product was performed on 1.5% agarose gel in tris-acetate EDTA (TAE) buffer, stained with ethidium bromide (50 l/l) and visualized on a UV- transilluminator. The size of the PCR products was determined using a 100 bp DNA ladder Figure (1).
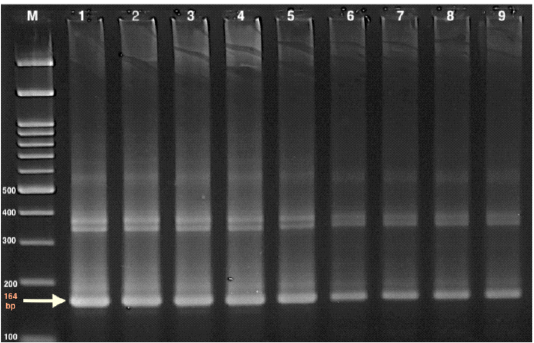
Figure 1: Gel electrophoresis of PCR product of TBR (164bp), for T. evansi using 1.5 % agarose gel in Tris-acetate EDTA (TAE) buffer Marker (M): 100 bp ladder marker 1: Control +ve 2-9: samples +ve
Statistical Analysis
The agreement among tests was measured using the kappa (κ) coefficient, values of κ coefficient 0–0.2 represented a slight agreement, 0.21–0.4 a fair, 0.41–0.60 a moderate, 0.61–0.8 a substantial, and >0.8 almost perfect agreement (Viera and Garrett, 2005). For each diagnostic test, sensitivity, specificity, the true and false positive predictive value, as well as the true and false negative predictive value were detected using crosstabs. Statistical software (SPSS Inc., Chicago, IL USA) has been used to conduct all statistical analysis.
Results
Clinical findings
Out of the 92 camels participating in the study, 36 were apparently healthy and 56 were clinically suspected experiencing some of the clinical signs suggesting T evansi infection, such as emaciation, powerlessness, conjunctival mucous membrane changes either paleness or congestion, superficial lymph nodes augmentation, edema of distal extremities Figure (2). Out of these clinically suspected cases, 34 gave positive by PCR and the most reported clinical findings in such 34 clinically affected camels were illustrated in Table (1).

Figure 2: Some important clinical signs noticed in examined camels; foot edema (a) and emaciation (b).
Prevalence of T.evansi
The overall prevalence of T evansi was 9.8 % (9/92) among the 92 camels screened with GSBS, 16.3% (15/92) with Ag-ELISA, and 58.7% (54/92) with PCR (Table 2).
The prevalence of T. evansi within the 56 clinically suspected camels was 16.1% (9/56) with GSBS, 26.8% (15/56) with Ag-ELISA and 60.7 % (34/56) with PCR, while, the prevalence in the apparent health cases was 55.6% (20/36) with PCR, but, no recorded cases using GSBS and Ag-ELISA. The difference in the prevalence between clinically suspected and apparently healthy animals was significant(x2= 6.41, df =1, P=0.011) with GSBS. While, was highly significant (x2= 11.52, df =1, P=0.001)
Table 1: Distribution of clinical signs in clinically suspected and affected camels
| Clinical signs |
Apparently healthy camels (n=36) |
Clinically suspected camels (n=56) |
Clinically affected camels* (n=34) |
|
| No. of animals (%) | No. of animals (%) | No. of animals (%) | ||
| Respiratory rate/minute | 10-15 | 36 (100) | 41(73.2) | 23(67.6) |
|
>15 |
0 | 15(26.8) | 11(32.4) | |
| <10 | 0 | 0 | 0 | |
|
Heart rate
|
35-40 | 36 (100) | 41(73.2) | 23(67.6) |
| >40 | 0 | 15 (26.8) | 11(32.4) | |
| <35 | 0 | 0 | 0 | |
|
Conjunctival mucous membrane
|
Bright rose red | 36 (100) | 10 (17.9) | 5 (14.7) |
| Congested | 0 | 10 (17.9) | 4 (11.8 ) | |
| Pale | 0 | 36 (64.3) | 25 (73.5) | |
|
Lymph nodes
|
Normal sized | 36 (100) | 44 (78.6) | 26 (76.5) |
| Enlarged | 0 | 12 (21.4) | 8 (23.5) | |
| Smaller | 0 | 0 | 0 | |
| Inappetance | 0 | 16 (28.6) | 3 (8.8) | |
|
Depression |
0 | 8 (14.3) | 4 (11.8) | |
| Recumbency | 0 | 1 (1.8) | 1(2.9) | |
|
Faecal abnormalities
|
Negative | 36 (100) | 48 (85.7) | 31(91.2) |
| Scanty feces | 0 | 7 (12.5) | 3 (8.8) | |
| Diarrhea | 0 | 1(1.8) | 0 | |
| Thigh muscles | Normal sized | 36 (100) | 10 (17.9) | 6 (17.6) |
| Thin | 0 | 46 (82.1) |
28 (82.4) |
|
(*): Clinically affected camels are those which were clinically suspected and confirmed positive by PCR
Table 2: Prevalence of Trypanosoma evansi and accuracy of GSBS, antigen-ELISA, and PCR for diagnosis of trypanosomiasis in camels.
| Test | GSBS | Ag-ELISA | PCR |
| No. (%) | No. (%) | No. (%) | |
| Positive | 9 (9.8) | 15(16.3) | 54 (58.7) |
| Negative | 83 (90.2) | 77 (83.7) | 38 (41.3) |
| PPV* | 100% | 100% | 100% |
| NPV** | 45.8% | 49.4% | 100% |
| Sensitivity | 16.7% | 27.8% | 100% |
| Specificity | 100% | 100% | 100% |
| Accuracy | 51.1% | 57.6 % | 100% |
| Prevalence | 9.8% | 16.3% |
58.7 % |
*PPV=positive predictive value, **NPV=negative predictive value.
Table 3: Kappa coefficient concordance test results among GSBS, Ag-ELISA, and PCR.
| GSBS | Ag-ELISA | |||||||||
| Κ | S.E. | 95.0% C.I | P | κ | S.E | 95.0% C.I | P | |||
| Lower | Upper | Lower | Upper | |||||||
| Ag-ELISA | 0.715 | 0.108 | 0.638-0.792 | 0.00 | ||||||
| PCR | 0.142 | 0.047 | 0.135-0.149 | 0.008 | 0.241 | 0.060 | 0.227- 0.255 | 0.00 | ||
with Ag-ELISA and with PCR (x2 = 34.67, df =1, P=0.00).
Regarding the prevalence of T. evansi among different age groups, in camels aged 1-3 years was 17.6% (3/17) with GSBS, 29.4% (5/17)with Ag-ELISA, and 52.9% (9/17) with PCR, in camels aged >3to7 years was 13.0 % (6/46) with GSBS, 19.6 % (9/46) with Ag-ELISA, and 63.0 % (29/46) with PCR, in camels >7 years was 0% (0/29) with GSBS, 3.4% (1/29) with Ag-ELISA, and 55.2 % (16/29) with PCR. The difference in the prevalence was significant (x2= 5.48, df =1, P=0.019) between 1-3 years and >7 years age groups, with GSBS. With Ag-ELISA, the difference in the prevalence was significant (x2= 8.43, df =1, P= 0.004) between 1-3 years and >7years age groups. On the other hand, no significant differences in the prevalence of T. evansi were recorded among different age groups with PCR.
The degree of agreement between the different diagnostic tests
Results of the Kappa agreement among the various diagnostic tests are reported in Table (3). A substantial agreement (k=0.715) between GSBS and antigen-ELISA was detected, while the agreement between GSBS and PCR was slight (k=0.142) and fair (k=0.241) between Ag-ELISA and PCR.
DISCUSSION
Several researchers have reported that camel trypanosomiasis is an enzootic disease in Egypt (Saleh et al., 2009; Sobhy et al., 2017). Many non-pathognomic clinical signs may be expressed in diseased camels; therefore, laboratory confirmation of the disease is necessary. Despite the poor sensitivity of many parasitological methods, these methods are widely used since they are inexpensive. Serological diagnosis of the disease may be conducted through antigen detection which is far more likely than antibody detection, in trypanosomiasis-enzootic areas since antibodies can persist for several months post-infection (Yadav et al., 2014). Besides, following therapy for surra, the animal is free from infection but still has detectable antibodies for a long period afterwards (Sengupta et al., 2019). Recently, an accurate diagnosis of T .evansi can be achieved using a molecular technique based on the detection of trypanosomal DNA by PCRs (Gutierrez et al., 2004). Many primers sets have been used for molecular diagnosis of T. evansi, of which TBR1/2 is the most sensitive one that also produced no cross-reaction with other pathogens (Pruvot et al., 2010). Therefore in this study, the PCR test using TBR1/2 primer was considered as the gold standard. In this study, the diagnostic performance of GSBS, Ag-ELISA, and PCR were estimated through the detection of the prevalence of camel trypanosomiasis in the Aswan governorate using these methods.
The most recorded clinical findings of surra in this study included pale mucous membranes of the conjunctiva, enlarged lymph nodes, thin muscles of thigh, and depression. Similar findings have been previously reported (Chaudhary and Iqbal, 2000; Rami et al., 2003) in camels affected by surra. In the current study, 22 cases were clinically suspected and showed clinical abnormalities, proved to be negative to T.evansi infection using the PCR technique, this result may be attributed to, the unspecific clinical signs of the infection of T. evansi that may confuse with other chronic debilitating diseases (Eyob and Matios, 2013). Since surra clinical signs may resemble those of many other diseases, therefore, the diagnosis of the disease must be confirmed by laboratory methods.
Concerning the diagnostic accuracy of different diagnostic methods, GSBS revealed 9 positive cases out of 92 camels with an overall prevalence rate (9.8%). This finding is in concurrence with the preceding study (Elhaig et al., 2013) as they recorded prevalence rate about (12%). But, lower rates were reported in camels in Egypt by (Abdel-Rady, 2008) (4.1 %), and (Abd-Elmaleck et al., 2014) (3.06 %). Differences in the prevalence of T.evansi with GSBS may be attributed to many factors such as, the low sensitivity of parasitological diagnosis of T. evansi in the subacute or chronic form of the disease, the number of examined camels or, the variable intensity of mechanical vectors that may be affected by seasonal variations (Abou-El-Naga and Barghash, 2016). Compared to the PCR, the sensitivity of GSBS was low (16.7%) since it was only able to detect 9 positive cases out of 54 PCR-positive cases, while the specificity of GSBS was 100%. This result is in agreement with (Muieed et al., 2011) they concluded that the parasitological methods used for the detection of trypanosomes are highly specific, but their sensitivity is relatively poor. (Elhaig et al., 2013) explained this low sensitivity as a result of low parasitemia in early or chronic infections. Therefore, the detected parasite prevalence using GSBS is lower than the true prevalence. Consequently, a more sensitive diagnostic method may be needed.
With antigen-ELISA, the overall prevalence of T.evansi was (16.3 %). This finding is comparable to that previously recorded in camels by (Omer et al., 1998) (13.8 %). Compared to PCR, the sensitivity of Ag-ELISA was low (27.8%), since it was only able to detect 15 positive cases out of 54 PCR- positive cases. The reported poor sensitivity of Ag– ELISA in the current study is in agreement with that previously reported by (Olaho-Mukani et al., 1993) who found that detection of either trypanosomal antigen or antibody showed poor results. The poor sensitivity of Ag-ELISA was explained by (Tehseen et al., 2015) as a consequence of fluctuating parasitemia and the low number of the parasite in chronic infections, as well as the incidence of antigen-antibody complexes. On the contrary, (Sengupta et al., 2019) reported a 97.4% sensitivity and 96.4% specificity of Ag-ELISA in detecting T. evansi Antigen in cattle.
The overall molecular prevalence (58.7 %) of T. evansi infection reported in this study is in parallel with that reported in camels in Egypt by (Abdel-Rady, 2008) (56.9 %). However, (Barghash et al., 2014) recorded a lower prevalence (46%) in camels in Egypt and in Algeria (13-13.6%)(Boushaki et al., 2019). These discrepancies in the prevalence rate may be attributed to geographic and climatic conditions and the presence of transporting vectors.
In this study, the prevalence that detected using all diagnostic methods was higher in clinically suspected cases than in apparently healthy animals. This result is coincided with (Elhaig et al., 2016). They reported that T.evansi is more prevalent in symptomatic camels. As regards, the effect of age on T.evansi prevalence, the non-significant difference reported in this study between different age groups using PCR is consistent with (Eyob and Matios, 2013). They concluded that all camels’ age groups are similarly exposed and infected with T.evansi.
To compare GSBS and Ag-ELISA to PCR, the prevalence rate of T. evansi using GSBS, antigen- ELISA, and PCR was, 9.8%, 16.3 %, and 58.7%, respectively. According to the kappa coefficient, the poor agreement (k=0.142) found in this study between GSBS and PCR is in concordance with that previously reported in camels by (Tehseen et al., 2015), they attributed that particularly in chronic cases due to low levels of parasitemia. In addition, (Gutierrez et al., 2004) concluded that in many hosts, PCR was more sensitive than traditional parasitological methods and has the advantage of being able to recognize parasites at the level of the organisms. The substantial agreement (k=0.715) that reported in this study between GSBS and Ag-ELISA can be clarified as both tests are useful methods for the diagnosis of acute trypanosomiasis (Singh and Singla, 2013). However, (Waitumbi and Nantulya, 1993) concluded that antigens detection of trypanosomes is more accurate than parasite detection. The fair agreement (k =0.241) that detected between Ag-ELISA and PCR may be related to false-negative results of Ag-ELISA in the early stage of infections (Thammasart et al., 2001).
In conclusion, the findings of this study revealed that both Giemsa-stain blood smears and antigen-ELISA had low sensitivity relative to PCR for the diagnosis of T.evansi in camels. Giemsa-stain blood smears can be used as a preliminary screening test for acute infection of T. evansi despite its low sensitivity relative to Ag-EISA since it is an inexpensive test. Moreover, the polymerase chain reaction can be used as a final confirmatory test.
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical disclosure
Informed consent form for the study have been approved and signed by camel owners. In this study, all animal procedures complied with institutional requirements guidelines obtained from the Ethics Committee of Assiut University, Egypt.
Author contribution
Arafat Sadek designed the experiment and supervised the laboratory work.
Khaled A.S. El-Khabaz designed the experiment, performed laboratory diagnosis, analyzed the results, wrote and revised the manuscript.
Sherief M. El-Genedy collected the samples, performed laboratory diagnosis and wrote the manuscript.
Magdy M. El-Gioushy performed the statistical analysis, wrote and revised the manuscript.
All authors read and finally approved the manuscript.
References





