Advances in Animal and Veterinary Sciences
Review Article
Studies on Multidrug-resistance Bacteria in Ruminants with Special Interest on Antimicrobial Resistances Genes
Abeer Mostafa Abdalhamed, Alaa Abdelmoneam Ghazy, Gamil Sayed Gamil Zeedan*
Department of Parasitology and Animal Diseases, National Research Centre, 33 Bohouth Street, Dokki, 12622, Giza, Egypt.
Abstract | Multidrug-resistant bacteria is a global problem in ruminants which resulting from inappropriate usage of antibiotics. Bacterial resistance genes can be transmitted transmit among ruminants, human, and the environment through food consumption (meat, milk and milk byproducts), either direct or indirect contact. Antimicrobial drug reduced entry; this strategy is being observed in Pseudomonas aeruginosa (P. aeruginosa) and Klebsiella spp reduced entry of beta lactam antibiotic, and vancomycin intermediate resistant S. aureus (VISA) strains. Activation of efflux mechanism to expel antibiotics from bacterial cell, efflux pumps mechanism is extruded antibiotics like tetracycline, macrolides, lincosamide, and streptogramins. Enzymatic degradation that leading to modification of antimicrobials either inside or outside the bacterial cell, as hydrolytic degradation of the β–lactam ring in penicillin and cephalosporin by bacterial β–lactamases. Modification of antimicrobial drug targets within the bacterial cell has been observed in methicillin resistant S. aureus (MRSA) through change or acquisition of different penicillin binding proteins (PBPs), and in vancomycin resistant Enterococcus (VRE). Other previously susceptible species may acquire resistance by genetic alterations within their genome, through mutations and /or horizontal gene transfer. In general, transfer of multidrug-resistance occurs through the processes of transduction (via bacteriophages), conjugation (via plasmids), and transformation through incorporation of free DNA segment into the chromosome. The present review is a preliminary study aimed to highlights on multidrug-resistant (MDR) bacteria problems in human and ruminants
Keywords | Klebsiella pneumoniae, Metaphylaxis, Multidrug resistant, Prophylaxis, Resistant gene, Ruminants, Staphylococcus aureus
Received | February 12, 2021; Accepted | March 12, 2021; Published | May 01, 2021
*Correspondence | Gamil Sayed Gamil Zeedan, Department of Parasitology and Animal Diseases, National Research Centre, 33 Bohouth Street, Dokki, 12622, Giza, Egypt; Email: gamilzee@yahoo.com
Citation | Abdalhamed AM, Ghazy AA, Zeedan GSG (2021). Studies on multidrug-resistance bacteria in ruminants with special interest on antimicrobial resistances genes. Adv. Anim. Vet. Sci. 9(6): 835-844.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.6.835.844
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Zeedan et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
The emergence of multidrug-resistant (MDR) bacteria are increased worldwide where most common antimicrobials are no longer effective for controlling the infectious diseases (Martinez and Baquero, 2000). The use of antimicrobials has been applied in animals for prevention, controlling, treating infection and improving growth rate (Portis et al., 2012; Ferri et al., 2017). Antimicrobial usage (AMU) such as amoxicillin, amoxicillin, quinolones, gentamicin, novobiocin, tylosin, tilmicosin, penicillin, erythromycin and tetracycline in ruminates (cattle, buffaloes, sheep, and goats) are used for meat-producing, and dairy animals for the treatment and diseases prevention (Portis et al., 2012; Garcia-Migura et al., 2014). The inappropriate use (overuse, misuse, and underuse) of antibiotics led to emerge of bacterial resistance to therapeutic dose. Increasing rate of antimicrobial-resistant (AMR) bacteria due to the development of new mechanism of resistance (Sengupta et al., 2013). The multidrug-resistance takes place as of the accumulation of different antimicrobials inside bacterial strain, which able to live in the presence of antimicrobial drugs so that standard treatments become ineffective or it required a longer time of treatment and causing high mortalities in animals (Munita and Arias, 2016). The antibiotic-resistant bacteria in animals initiated after the first use of antibiotics for instance Staphylococcus aureus (S. aureus) was found to be resistant to penicillin, followed by Escherichia coli (E. coli), Shigella spp, and Salmonella enterica (S. enterica) as well as vancomycin-resistant Enterococcus spp (VRE), methicillin-resistant S. aureus (MRSA), and multidrug-resistant Acinetobacter baumannii (MDR A. baumannii) (Cantas et al., 2013). There are many factors relating to the occurrence of AMR strains as the antimicrobial itself (dosage, frequency, selection pressure for unblocking gene expression) resulting in the development of resistance genes promoting occurrence of mutations in genes that generating resistant organism, in ruminants about 75 to 90 % of antimicrobials which used are excreted un-metabolized which increases the antimicrobial residues in farm environments (Martinez, 2009; Ateba Ngoa et al., 2012). Also, resistant in microbes can build up through mutating existing genes (vertical) (Martinez and Baquero, 2000), acquiring a new gene from environment or through horizontal gene transfer (HGT) (Heuer and Smalla, 2007; Von Wintersdorff et al., 2016). All pathogenic and commensals resistant bacteria can be transmitted from ruminants to human through consumption of meat and milk, or by direct or indirect contact with animals or their waste (O’Connell, 2013; Rodriguez-Mozaz et al., 2015). AMR genes and bacteria can be transmitted to long distant via airborne from large cattle feedlots in semi-arid areas (McEachran et al., 2015; Alhaji et al., 2018). Manures contaminated with resistant bacteria are transmitting resistance to the surrounding environment and contaminated lands through transfer resistance genes between different animals and the environment (Pehrsson et al., 2016; Rousham et al., 2018). Therefore, the present review is considered as a preliminary study aimed to throw light on the problem of multidrug-resistant bacteria in ruminants.
Antibiotic Resistance History
Introduction of the first antimicrobial sulfonamides in 1937, and specific resistance has been reported (Davies, 2006). Since Alexander Fleming discovered the first antibiotic, penicillin, after that several antibiotics have been discovered as tetracycline, chloramphenicol and gentamicin, unfortunately, a resistance phenomenon was found in S. aureus, it was no longer susceptible to penicillin (Abraham and Chain, 1940; Davies and Davies, 2010). Followed by Streptococcus pneumoniae in 1967, and Enterococcus faecium (E. farcium) in 1983 joined to the list of bacteria resistant to penicillin (Lewis et al., 1995), after discovering streptomycin in 1944 for the treatment of tuberculosis (TB). However, mutant strains of Mycobacterium tuberculosis resistant to therapeutic dose was found (Kong et al., 2010; Baquero et al., 2011; Bhullar et al., 2012).
Multidrug-resistance was first found in enteric bacteria such as Escherichia coli (E. coli) and Salmonella spp in 1950 - 1960 (Spang et al., 2013). The unexpected identification of genetically antibiotic resistance in Japan in the mid-1950, by introducing a genetic concept as antibiotic resistance genes could be disseminated by bacterial conjugation throughout an entire species of bacterial pathogens (Ezeamagu, 2014; Munita and Arias, 2016). All classes of antimicrobials including sulfonamides, penicillin, tetracycline, ampicillin, aminoglycosides, and cephalosporin (cephalexin) that used in therapeutics purpose in veterinary and human medicine rise drug resistance as shown in Figure (1) (Berendonk et al., 2015; Collignon et al., 2016).
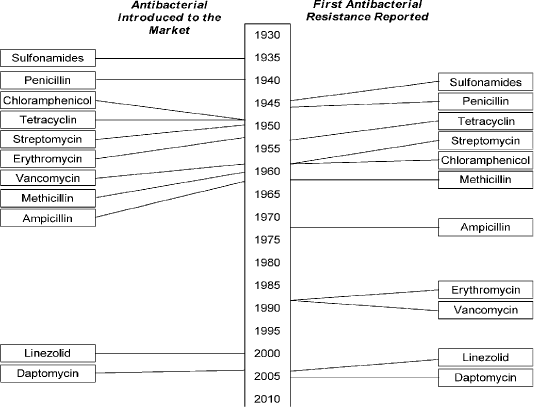
Figure 1: Showed that the timeline of introduction of antibacterial and resistance development . (Holmes et al., 2016). https://www.amstewardship.ca/factsheet/veterinarians/antimicrobial-resistance-
Status of antimicrobial resistance in Egypt
There are several reports about the status of AMR in ruminants in Egypt (Ahmed, and Shimamoto, 2011; Abdalhamed et al., 2018) and Ahmed and Shimamoto (2011) reported the genetic basis of MDR bacteria isolated from bovine mastitis in Egypt, and found that 30.4% Gram-negative bacterial isolates possess at least one antimicrobial resistance gene, and found the most prevalent species were Enterobacter cloacae (8 isolates, 7.1%), Klebsiella pneumoniae (K. pneumoniae) (6.3%), Klebsiella oxytoca (K. oxytoca) (6.3%), E. coli (4.5%), and Citrobacter freundii (2.7%). The first report about the high prevalence of multi-resistant extended spectrum beta-lactamases (ESBLs) producing E. coli recorded in cattle (Pfeffer et al., 2016). Romera et al. (2014) isolated the CMY-, CTX-M-, OXA-, SHV-, and TEM-β-lactamases genes in E. coli and Salmonella spp. from calf’s diarrhea. Tarabees et al. (2016) recorded the most common MDR bacteria causing pneumo-enteritis in small ruminants in three Egyptian provinces; E. coli was the highest percentage followed by S. aureus and Salmonella (70.99%, 5.34% and 3.82%, respectively) as detected by PCR. Abdalhamed et al. (2018) isolated and identified AMR S. aureus, coagulase-negative staphylococci (CNS), E.coli, Streptococcus spp., Klebsiella spp. and Pseudomonas spp from sheep and goat mastitic milk. Zeedan et al. (2018) recorded that MDR of E. coli was 18.7%, S. aureus was 7.25% and Salmonella spp was 16.12% isolated from bovine calf diarrhea respectively at different governorates in Egypt. Aziz et al. (2018) determined prevalence of Salmonella spp in 90 calves’ feces in Beni-Suef governorate, was 5.6%. The predominant serovar was Salmonella kentucky and added that 100% of the isolates were MDR. The existence of class 1 integron and gene cassettes in the resistant isolates were bla SHV and bla TEM for β-lactams, aadA2 for aminoglycosides (streptomycin), tetA and tetB for tetracycline, dfrA and mphA for macrolides.
In cattle, buffaloes, sheep and goat, as amoxicillin, penicillin, erythromycin, quinolones, gentamicin, novobiocin, tylosin, and tetracycline are extensively used. In meat-producing animals, antibiotics are mainly used for the treatment and prevention of shipping fever, mastitis, lameness, respiratory diseases and gastrointestinal disorders (Arsenault and Kogut, 2016), which are the most common problems in sheep and goats, it is preferable to administer antimicrobials in other ways than the oral route (feed or water), with the exception of certain sulfonamides and tetracycline which can be absorbed efficiently by the rumen (Arsenault and Kogut, 2016). As a result of misuse of antimicrobial drugs that driving force that developing AMR in ruminants in both commensal and pathogenic bacteria (Pehrsson et al., 2013). Emergence of AMR strains in ruminant is related to 75 to 90 % of antimicrobials where it is excreted, mostly un-metabolized which increasing concentration of drug residues in farm environments (Martinez, 2009). Resistant bacteria can be transmitted through food consumption (meat and milk) from ruminants to humans, or through direct or indirect contact from animals or their waste in the environment (Hassell et al., 2019) as shown in Figure (2)
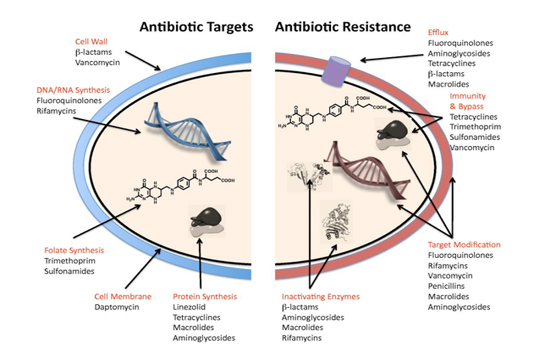
Antibiotics target and mechanisms of resistance
Antibacterial agents act on bacterial cell wall by targeting essential processes such as inhibiting their cell-wall construction, disrupting the structure and function of their cell membrane, preventing the synthesis of vital proteins or interfering with synthesis of genomic RNA or DNA as shown in Figure (3) (O’Connell et al., 2013).
Some of these agents inhibit cell-wall construction leads to bacterial cell death, termed bactericidal. Other agents, such as the tetracycline, which inhibit protein synthesis, referred as bacteriostatic, they simply prevent the growth of the bacteria. Some antibacterial agents effect is against a narrow spectrum of bacteria, where glycopeptides only display activity against Gram-positive organisms, whereas other antibacterial, as β-lactams, target processes across different species and classified “ broad-spectrum antibacterial agents” (Luc, 2015). Although, bacterial species are not susceptible to all antibiotics; some species are intrinsically resistant i.e. natural resistance. Such resistance can be due to a specific protective mechanism against the antibiotic, or due to genetic trait with a different function, that happens to also convey resistance (Munita and Arias, 2016).
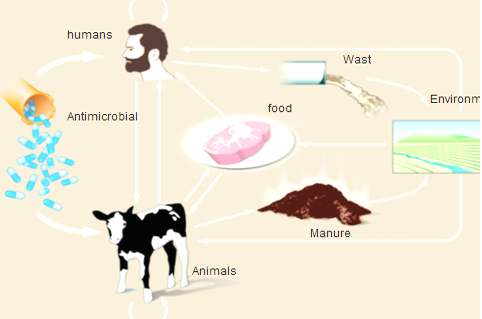
Figure 2: Routes of transmission of drug-resistant between different farm animals, environment and Humans (Hassell et al., 2019).
The mechanisms of bacterial resistance for different class of antimicrobials have been summarized in Figure (3). These mechanisms can be organized into four categories; I: reduction of entry or access to the target site of antimicrobial drug has been observed in Pseudomonas aeruginosa (P. aeruginosa), Klebsiella spp resist to beta–lactam antibiotic, and vancomycin intermediate resistant S. aureus strains which have thickened cell wall to trap the drug before entry into the cell (Munita and Arias, 2016). II: Activation of efflux mechanism to expel antimicrobial agents from the bacterial cell, like tetracycline, macrolides, lincosamide, and streptogramins. Many other pumps possess ability to expel multiple drugs which can expel a variety of structurally different antimicrobials (Sanchez et al., 2016). This strategy has been observed in E. coli and other Enterobacteriaceae against tetracycline and chloramphenicol, in S. aureus and Streptococcus pneumonia against fluoroquinolones (Marr et al., 2006; Munita and Arias, 2016). III: Enzymatic degradation or modification of antimicrobials either inside or outside of the bacterial cell, a classic example is the hydrolytic degradation of the β–lactam ring in penicillin and cephalosporin by the bacterial β–lactamases (King et al., 2016). IV: Modification of antimicrobial drug targets within the bacterial cell observed in methicillin resistant S. aureus (MRSA) through change or acquisition of different penicillin binding proteins, and in vancomycin resistant Enterococcus (Blair et al., 2015). Other susceptible species may acquire resistance by genetic alterations within their genome, either by mutations or horizontal gene transfer (Von Wintersdorff et al., 2016). In general, transfer of resistance determinants occurs through the processes of transduction via (bacteriophages), conjugation via (plasmids), and transformation through incorporation of free DNA segment into the chromosome as in Figure (4).
Acquired resistance as chromosomal mutations in Mycobacterium tuberculosis leading to rifampicin resistance, fluoroquinolone resistance due to mutation in the drug’s targets DNA gyrase and topoisomerase IV, and horizontal acquisition of mecA in methicillin resistance (Bajaj et al., 2016). Intrinsic and acquired resistance can affect the four mentioned major resistance pathways. Naturally, occurring efflux pumps in P. aeruginosa can be overexpressed by mutations in repressor genes, leading to increased resistance to those antibiotics that act as substrates for the respective pump, the transmission of resistance genes from bacteria in livestock animals (Pang et al., 2018).
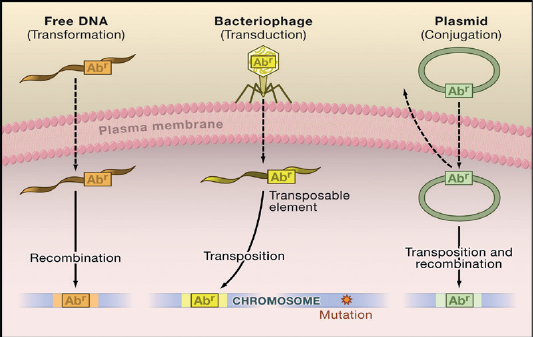
Figure 4: Antibiotic-resistant bacteria that are caused by a mutation in the target gene in the chromosome (Arcilla et al., 2016).
Transmission of chromosomal AmpC genes to plasmid; the plasmid-mediated spread of quinolone resistance (Halaby, 2018). In addition, plasmid carries genes that encode resistance to different classes of antibiotics, leading to transfer and spread of multidrug resistances (Zarei-Baygi et al., 2019).
Antibiotics resistance genes
Microbes can build up resistance to antibiotics through mutating existing genes (vertical) (Martinez and Baquero, 2000) or through acquiring a new gene from environment and other species by horizontal gene transfer (HGT) (Heuer and Smalla, 2007; Von Wintersdorff et al., 2016). HGT can occur between Gram-negatives and Gram-positives; aerobes and anaerobes; and non-pathogenic bacteria in human or animal (Pérez Gaudio, 2018). The sharing of genetic materials between bacteria occurs mainly through mobile genetic element (Wellington et al., 2013). Small amount of antimicrobials concentrations could enhance the survival of gene mutations in a bacterial population. Genetically, bacteria have evolved to be diploid division, which help them to express and survive both susceptibility and resistant traits mechanisms e.g. efflux pumps in cell membranes can be adopted by bacteria as observed E.coli strains resistant (Zhang et al., 2016; Sharma et al., 2018; Frenoy and Bonhoeffer, 2018; Asante and Osei Sekyere, 2019). It has been observed that the resistance among bacterial species through antibiotic-resistant genes and the major genes leading to AMR includes blaTEM genes blaTEM- transposons in commensal E. coli for the antibiotics penicillin/amoxicillin/ampicillin (Neyra et al., 2014), or via erm gene cluster for macrolides (erythromycin/tylosin/tilmicosin/ kitasamycin/ oleandomycin) of AMR gene and vanA-enterococci from cattle and sheep (Sharma et al., 2014).
Rojas-Lopez et al. (2018) explained E. coli virulence genes transmitted through fecal contamination of milk products are source for human outbreaks association with MDR Shiga-like toxin (stx1 and stx2) genes. Aziz et al. (2018) identified integrons class 1 in MDR Salmonella isolates from neonatal calf diarrhea. The identified genes class 1 integrons were (aadA1, aadA2 and aadA5) for aminoglycoside adenylyl transferase type A that conferring resistance to streptomycin, spectinomycin, and dihydrofolate reductase genes (dfrA1, dfrA15 and dfrA15), conferred resistance to trimethoprim. Xiong et al. (2018) reported that the food-producing animals may be play a role as primary reservoir of resistant pathogenic strains, and transfer resistance genes between animals, humans, and the environment. Asante and Osei Sekyere (2019) documented that the resistance genes and mobile genetic elements (MGEs) were IS16 and Tn916 highly associated with erm (B) and tet (M) in Enterococcus faecium (E. faecium), ST18, ST80 and ST910 in Streptococcus agalactiae (S. agalactiae), ST612, ST616 and ST617 in Enterococcus faecalis (E. faecalis), and in Streptococcus (S. pyogenes) were emm18, emm42, emm76 and emm118, while SCCmec associated with mecA in S. aureus, and ST5, ST80, ST8, and ST88 were in S. haemolyticus, and the antibiotic resistant rate were found in humans, animals and environment such as methicillin-resistant of S. aureus (MRSA) for beta-lactams (blaZ, mecA) and macrolides (msr(A)/msr(B),was isolated from calves, lambs and goats in Spain (Ruiz-Ripa et al., 2019).
Taghadosi et al. (2019) investigated the prevalence of antibiotic resistance, class 1 and 2 integrons in Extended Spectrum β-Lactamases (ESBL) genes of pathotypes E.coli isolated from cattle and goats in Iran. Tian et al. (2019) investigated virulence genes of Streptococcus in 735 mastitic cow’s milk in China and found that the highest resistance rate to tetracycline (98.44%) and oxacillin (98.44%), followed by penicillin G (96.88%) and doxycycline (96.88%), and the lowest resistance was observed with ciprofloxacin (1.56%), seven virulence genes detected in 59 (92.19%) isolates harbored at least one gene, twenty-four classes of gene patterns found in the isolates and the patterns of bca (9.38%) and cfb (9.38%) for Streptococcus isolates.
Xia et al. (2019) revealed genotyping of Campylobacter jejuni (C. jejuni ) sequence type 2862 (ST2862) and Campylobacter coli (C. coli) ST902 were the predominant genotypes in sheep. Multiple antibiotics-resistant bacteria emerging in dairy cows’ mastitis as a result of extensive/uncontrolled drug use, based therapy, horizontal gene transfer, and/or spontaneous genetic mutations have increased health risk to humans by contaminating milk and milk products.
Multidrug-resistant bacteria in ruminants
Presence of resistant pathogenic strains in dairy cattle, calf, beef, sheep, goats and feedlot animals lead to develop MDR and the primary reservoir of pathogenic organisms which increase morbidity and mortality (Beceiro et al., 2013) as in MDR Gram-negative, and Gram-positive bacteria (Fair and Tor, 2014; Holmes et al., 2016).
MDR-Gram negative bacteria
Escherichia coli: E.coli is a commensal bacteria in the gastrointestinal tract of healthy animals, as in calves, goats and sheep, However some types of E. coli, can cause intestinal infection due to acquire certain virulence factors and antimicrobial resistant gens (Beceiro et al., 2013; Miró-Canturri et al., 2019). Genetic investigations of antimicrobial resistance in healthy lactating dairy cows have found that E. coli is an important reservoir for tetracycline and other antimicrobial resistance determinants (Landers et al., 2012; Takada et al., 2016). Cattle are the most common farm animals have E. coli O157:H7 and water buffaloes act as carriers (Catford et al., 2014). Cow calves have lower risk than feedlots animals harbor E. coli resistant to tetracycline, sulfamethoxazole, and streptomycin and have no resistances to ceftriaxone or ciprofloxacin and less than 1% of isolates were resistant to gentamicin, nalidixic acid, and ceftiofur (Barlow et al., 2015). E. coli isolates were resistant to beta-lactams such as penicillin (Sartelli et al., 2016). Furthermore, multiple antibiotic resistant E. coli strains have also been isolated from milk (Gonzalez-Escalona et al., 2016). Argud et al. (2017) reported plasmid-mediated mcr-1 gene co-resistant to β-lactam, florfenicol, and fluoroquinolone antimicrobial compounds from 150 E. coli isolated from diarrhea and mastitic cattle in Europe and Asia. Also, Hille et al. (2017) found that the prevalence of Cefotaxime-resistant E. coli in dairy and beef cattle in Germany were 70% and 41%, respectively. Abdalhamed et al. (2018) found that E. coli was 8.3% from mastitis of sheep and goat, and found the resistance for different used antibiotics were 67.14%. Cheng et al. (2019) demonstrated prevalence AMR of E. coli and Klebsiella spp from bovine mastitis in Chinese dairy herds was high to amoxicillin/clavulanate potassium (81% and 38%, respectively), followed by tetracycline (only Klebsiella spp. 32%), and high proportion (27%) of isolates were multidrug resistant.
Salmonella spp: Salmonella infections are an important cause of mortality and morbidity in cattle (Gal-Mor et al., 2014). Salmonellosis is a common enteric bacterial infection of cattle and sheep, infection may occur at any age are more severe in calves from the first 2 weeks to 3 months of their life (Aviv et al., 2016). The emergence of MDR Salmonella serotypes are more virulent lead to an increase in mortality rates (Cragg and Newman, 2013). El-Sharkawy et al. (2017) compared between MDR Salmonella from calves and adult cattle and found that the percentage were 22.0% and 95 %, respectively. Nelson et al. (2019) reported that the multidrug-resistance Salmonella to tetracyclines, sulfonamides, streptomycin, kanamycin, chloramphenicol, and some of the β-lactam antibiotics (penicillins and cephalosporins). Demirci et al. (2019) examined 231 raw milk samples from cows, goats, sheep, and donkeys and found that 2.16% recorded positivity for MDR Salmonella spp. by multiplex real-time PCR.
Campylobacter spp: Campylobacter fetus (C. fetus) is commonly present in the gastrointestinal tract of cattle and sheep, it is associated with diarrhea (Emele et al., 2019). Enzootic infectious caused infertility in cattle infected by Campylobacter fetus subsp venerealis causes abortions in sheep, goats, and cattle, has been associated with C. fetus subsp. fetus and to a lesser extent with Campylobacter jejuni (C. jejuni) ( Xia et al., 2019). Han et al. (2019) evaluated drug resistance rates, of C. jejuni from cattle farms, slaughterhouses, and people had diarrhea for several antibiotics associated with resistance genes and they found high frequencies of resistance to tetracycline (100%), ciprofloxacin (95%), and nalidixic acid (86%), Also, low frequencies of resistance to florfenicol (0%), erythromycin (5%), and gentamicin (8%). Xia et al. (2019) examined Campylobacter profiles in sheep naturally infected with Campylobacter, medicated with tetracycline in feed, while the other received feed without antibiotics. And they found that the fecal and bile samples were positive for Campylobacter jejuni and Campylobacter coli, with no differences between the medicated and nonmedicated groups. All isolates were resistant to tetracycline. C. jejuni had low resistant to fluoroquinolone (FQ), where C. coli were 95.0% resistant to FQ.
MDR-Gram positive bacteria
Staphylococcus: S. aureus is part of the normal flora in dairy herds worldwide (Al-Ashmawy et al., 2016). Coagulase-negative staphylococci (CNS) are the most prevalent bacteria isolated from subclinical mastitis of sheep and goats (Addis et al., 2016). Holmes et al. (2016) isolated and characterized MRSA, oxacillin-susceptible mecA-positive S. aureus (OS-MRSA), and methicillin-susceptible S. aureus (MSSA) from milk samples of bovine mastitis and they reported that the Staphylococcus spp was 53%. 60 of isolates identified as S. aureus (98.4%) and 1 isolate was Staphylococcus epidermidis (1.6%). The presence of the mecA gene was 48.3% of S. aureus isolates, 23.3% of MRSA and 25.0% of OS-MRSA. Investigation of the prevalence of MRSA beta lactams group of bacteria, which include methicillin, oxacillin, penicillin and amoxicillin in small ruminants’ milk (Zeedan et al., 2014, Papadopoulos et al., 2018). Obaidat et al. (2018) determined the antimicrobial resistance of mecA and mecC MRSA in dairy cattle, sheep, and goat which exhibited resistance toward gentamicin, clindamycin, rifampicin, neomycin, fusidic acid, erythromycin, tetracycline, and ciprofloxacin Saei and Panahi, (2019) observed the resistance of S. aureus isolated from nasal swabs and mastitic milk samples of dairy animals (cows, sheep, and goats) using antimicrobial susceptibility testing (AST) for penicillin (64.2%, 43/67), followed by tetracycline (23.9%, 16/67), erythromycin (22.4%, 15/67), and streptomycin (17.9%, 12/67), respectively.
Enterococcus spp: Enterococci are commensal bacteria gastrointestinal (GI) tract of bovine and human. It causes mammary infection in sheep and goats (Underwood et al., 2015). E. faecalis infections associated with plasmid-encoded hemolysin (cytolysin) is responsible for pathogenesis during animal infection, and the cytolysin in combination with high-level gentamicin resistance increased five-fold causing death (Ali et al., 2017). E. faecalis is resistant for many antimicrobial agents as aminoglycosides, aztreonam, cephalosporins, clindamycin,penicillins, nafcillin, oxacillin,trimethoprim-sulfamethoxazole and vancomycin.
Rehman et al. (2018) found 96·7% of the examined dairy cows were positive for Enterococcus hirae, E. faecalis and E. faecium. The highest percentage of resistant isolates were to lincomycin (92·3%), flavomycin (71·9%) and tetracycline (24·5%), Multi-drug resistance ≥ 2 antimicrobials were observed for seven antimicrobials. Cheng et al. (2019) investigated AST of MDR enterococci isolated from bovine clinical and subclinical mastitis in China and found that subclinical mastitis was 34.3%, and clinical mastitis was 21.4%, more than 50% of the total isolates were resistant to penicillin, ceftiofur, tylosin, lincomycin, and oxytetracycline.
Conclusion
Antibiotics had been used in sheep, goats, cattle and buffaloes for treating, preventing, controlling of diseases and as growth promoters. All classes of antimicrobials that including sulfonamides, penicillin, tetracycline, ampicillin, aminoglycosides, and cephalosporin (cephalexin) used in veterinary and human medicine rise drug resistance. The resistance in bacteria is a specific protective mechanism against antibiotic, it can be due to a genetic trait as intrinsic resistance, or acquire resistance by genetic alterations within their genome, either by mutations or horizontal gene transfer through the processes of transduction, conjugation, and transformation. The genetic information transmitted between bacteria through mobile genetic element that includes phages, plasmids, and transposons. Once, resistance genes transferred to bacteria, its loss activity of the antibiotic. Methicillin-resistant S. aureus (MRSA) for beta-lactams (blaZ, mecA) and macrolides (msr(A)/msr(B) reported among the S. aureus isolates from calves, lambs and goats. In dairy herds, methicillin-resistant S. aureus (MRSA) was associated with clinical mastitis. Streptococcus isolates had the highest resistance rate to tetracycline, oxacillin, penicillin G and doxycycline. The emergence of MDR Salmonella serotypes are more virulent and lead to an increase in mortality rates. Salmonella has multidrug resistance to tetracyclines, sulfonamides, streptomycin, kanamycin, chloramphenicol, and some of the β-lactam antibiotics (penicillins and cephalosporins). Some types of E. coli, particularly E. coli O157:H7 causes intestinal infection as it acquiring certain virulence factors and antimicrobial resistant gens. Genetic investigations of antimicrobial resistance in lactating dairy cows have found that E. coli is an important reservoir for tetracycline and other antimicrobial resistance determinants. Multiple antibiotic resistant E. coli strains have been isolated from milk. E. coli isolates were resistant to beta-lactams such as penicillin. Acquired resistance as chromosomal mutations in TB lead to rifampicin resistance, fluoroquinolone resistance due to mutation in the drug’s targets DNA gyrase and topoisomerase IV, and horizontal acquisition of mecA in methicillin resistance.
Acknowledgements
The authors are thankful to National Research Centre, Dokki, Egypt, for facilities during this work.
COnflict of interest
The authors declared that they have no competing interests.
Authors’ Contribution
Abeer M. Abdalhamed & Gamil SG Zeedan, found research idea, and drafted the manuscript. Alaa A Ghazy sharing in the conception of the research idea, and helped in manuscript preparation.
References