Advances in Animal and Veterinary Sciences
Research Article
Molecular Comparison Between Resistance Genes in Staphylococcus aureus Clinically Isolated from Cattle and Camels in Southern Egypt
Alsagher O. Ali1, Waleed Younis2, Hassan Y.A.H. Mahmoud1*
1Division of Infectious Diseases, Animal Medicine department, Faculty of Veterinary Medicine, South Valley University, Qena 83523, Egypt; 2Departments of Microbiology, Faculty of Veterinary Medicine, South Valley University, Qena 83523, Egypt.
Abstract | S. aureus is one of the most common organisms associated with infections among dairy animals with frequent and rapid development of antibiotic resistance. The presence of genes encoding aminoglycoside resistance tetracycline resistance, and beta-lactam resistance were detected by whole genome sequencing. Seventy-nine S. aureus clinical isolates were isolated from milk samples of dairy cattle and she-camels in Southern Egypt. The sequences of resistant genes extracted from the whole genome sequences of the bacterial chromosome that was done through shotgun sequencing, through phylogenetic analysis, all resistant genes sequences were arranged into two main clusters. The first clusters contain mecA genes, aminoglycosides, fusB gene as well as blaz gene except A11, and two reference genes (AJ302698 and AP003139). The second cluster contains mainly the tet(k) and tet(38) genes which are more similar to each other, in addition to mecA reference gene (AB512767) and two blaz reference genes (AJ302698 & AP003139). The phylogenic tree revealed that all resistant genes are more or less stable genes such as mecA, aminoglycosides, tet(k), tet(38), and fusB except blaz gene showing more variations. Molecular detection continues to be increased to establish antimicrobial resistance trends for S. aureus found in subclinical mastitis of dairy animals.
Keywords | Antibiotic resistance, Camel, Cattle, S. aureus, Egypt
Received | October 10, 2020; Accepted | December 15, 2020; Published | January 15, 2021
*Correspondence | Hassan YAH Mohamoud, Division of infectious diseases, Animal Medicine department, Faculty of Veterinary Medicine, South Valley University, Qena 83523, Egypt; Email: mhassan@vet.svu.edu.eg
Citation | Ali AO, Younis W, Mahmoud HYAH (2021). Molecular comparison between resistance genes in S. aureus clinically isolated from cattle and camels in southern egypt. Adv. Anim. Vet. Sci. 9(3): 453-461.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.3.453.461
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Mahmoud et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Resistance to antimicrobial agents is regarded as one of the world’s main and increasingly global issues, especially among nosocomial pathogens. Staphylococci have been one of the most common causes of nosocomial infections. Multidrug-resistant staphylococci pose a rising alarm for public health. The rise of drug-resistant virulent strains of S. aureus, particularlymethacillin resistant S. aureus (MRSA) is a severe problem in the treatment and control of staphylococcal infections (Livermore, 2000; Zapun et al., 2008; Neamat-Allah and Hend, 2016). Staphylococci resistance to methicillin (MRSA) makes infections difficult to treat since they are resistant to most antibiotics such as beta-lactam, aminoglycosides, and macrolides. The recent series of numerous S. aureus gene and bioinformatics are highly promising in recognizing and characterizing target genes (Land et al., 2015; Everitt et al., 2014). It may also be inferred from the pathways by which genes have antibiotic resistance. These genes may also be used for a greater understanding of pathogenesis and pathogen; therefore, it is of interest to predict its mechanism by identifying the unique antibiotic-resistant genes from the genomes of different S. aureus strains (Everitt et al., 2014).
MecA, mecR1, and mecI are the main methicillin resistances proteins that have been identified and expressed by most strains. The genes were well studied as analyses in most S. aureus strains, including MRSA (Ito et al., 2003; Monecke et al., 2010; Stegger et al., 2012), with antibiotic-resistant genes. The mecR1 analysis of antibiotic-resistant genes plays a role in penicillin interactive as a potency anti-represent for methicillin resistance. Lowy has explored the various pathways by which these genes contribute to antibiotic resistance (Lowy, 2003). MecI and mecR1 of antibiotic-resistant genes regulate mecA reaction to β-lactam antibiotics in a method like the regulation of blaZ by blaR1 and blaI genes in penicillin sensitivity (Lowy, 2003; Stegger et al., 2012). The main mechanism for penicillin resistance is triggered by the development of an exogenous mecA gene, which encodes an extra penicillin-resistant β-lactam protein called PBP-2.
Aminoglycoside resistance mechanisms include decreased absorption, reduced cell permeability, ribosomal binding site change by mutation expression of rRNA methylases, and aminoglycoside modification enzyme production. The aacA-aphD gene resistance to all aminoglycosides, except streptomycin and neomycin; it is clinically used to encode the bifunctional enzyme amino glycoid-6-N-acetyltransferase/2-O-nucleotidytranferencease (van Den Braak et al., 1999; Landecker, 2016).
Aminoglycosides are broad range antibiotics that are used to treat S. aureus in combination with other antibiotics, such as ß- lactams (Ramirez and Tolmasky, 2010). The most common cause of aminoglycoside resistance is the non-activation of aminoglycoside antibiotics by aminoglycoside modified enzymes (such as aminoglycoside phosphotransferase, acetyltransferases, nuclear transferase enzymes). Aac(6’)-Ie-aph(2), «aph(3’)-IIIa and ant(4’)-Ia, which can be found on plasmid or chromosomes and often contain transposable components, are the typical aminoglycoside-modifying enzymes that encode S. aureus (Ramirez and Tolmasky, 2010; Fatholahzadeh, 2009; Emaneini et al., 2009).
Tetracyclines have been used to treat and prevent bacterial infection with a broad spectrum of antibiotics. (Ardic et al., 2005) Many tetracycline-resistant bacteria have developed tetracycline resistance genes. Staphylococcus aureus has two major resistance mechanisms: aggressive efflux arising from the acquisition of tetK- and tetL genes found in plasmid and ribosomal protection by elongation factors of tetM- or tetO (Esposito et al., 2009; McCallum et al., 2010). The topical agent for infection of the skin and certain systemic infections caused by S. aureus was fusidic acid (Besier et al., 2003). S. aureus has two main fusidic acid resistance mechanisms reported: altering the target drug caused by fusA (Nagaev et al., 2001; O’Neill et al., 2004; Norström et al., 2007) or rplF (Lannergård et al., 2009) mutations and defending the drug target site with fusB, fusC, and fusD (Ardic et al., 2005; Ramirez and Tolmasky, 2010). FusB, FusC and FusD were found in S. aureus and coagulase-negative staphylococci (McLaws et al., 2008; O’Neill and Chopra, 2006; O’Neill et al., 2007) and fusD was an intrinsic factor causing fusidic acid resistance in S. saprophyticus (O’Neill et al., 2006).
Materials and methods
A total 240 milk samples included (140 milk samples from dairy cattle and 100 milk samples from she-camels) were collected in sterile plastic tubes from the Red Sea and Qena governorates, Egypt. All animals were apparently healthy and they did not show any signs of clinical mastitis. The milk samples were directly transferred in icebox to the laboratory of infectious diseases, department of animal medicine, Faculty of Veterinary Medicine, South Valley University. Animals have been tested concerning the presence of subclinical mastitis. Clinical parameters including body temperature, heart rate, and respiratory rate were recorded. Animals with no local or systemic infection were considered healthy.
Isolation and culturing of Staphylococcus aureus
The obtained milk samples (10 µL) were inoculated onto 5% sheep blood agar (Oxoid) incubated at 37°C for 24 hours. The suspected colonies of S. aureus were subculture on Baired-Parker media (Oxoid). A few standard colonies (black colonies with clear zone around) were collected and picked up by a bacteriological loop, then immersed in nutrient glycerol broth stock and preserved for further identification at -80 ° C.
Microbiological data
All isolates were tested for different bacteriological assays to detect the most properties of S. aureus, such as bacterial sensitivity tests. Multiplex PCR used to classify S. aureus by using femB, mecA, and mecC genes into MRSA and MSSA. The data was published previously (Ali, 2015; Ali et al., 2015; Ali, 2016; Ali and El Sherief, 2016).
Biochemical tests
Gram staining, characteristic features of colonies on Baired-Parker media (Oxoid), hemolysis patterns on blood agar enriched with 5% (v/v) sheep blood, catalase and coagulase tests were used to identify S. aureus isolates. Two separate types of coagulase testing were used, namely the slide coagulase test and the tube coagulase test (Field and Smith, 1995).
Detection of resistant genes
The classification of S. aureus into methicillin-susceptible or resistant (MSSA or MRSA) was confirmed by using mecA gene. Multiplex PCR was used to ensure the presence of mecA gene, in addition to the mecC gene, which was detected recently as a homolog for mecA and the femB gene used as a confirmatory gene for S. aureus (Nakagawa et al., 2005; Paterson et al., 2014). For Detection of mecA, blaZ, aac(6’)- aph(2’’), aph(3’)-III, ant(6)-Ia, tet(K), tet(38), and fusB genes. The genes were extracted from the S. aureus genome obtained by shotgun sequencing (Sanger Institute, UK). mecA, blaZ, aac(6’)- aph(2’’), aph(3’)-III, ant(6)-Ia,, tet(K), tet(38), and fusB genes were extracted from the bacterial genome of local bacterial isolates (A1, A2, A3, A4, A5, A6, A7, A10, A11, A12, A13, A15). The following resistant genes (9 sequences) were retrieved from the NCBI GENEBANK and their accession numbers (M13771, AB512767, U38428, AY825285, M26832, AF330699, AB033763, AJ302698, AM292600, AP004832, AP003139, J01764, and FR821779).
Whole-genome sequencing
The DNA of The S. aureus both resistant and susceptible strains (MRSA and MSSA) were extracted and the genomic DNA used to obtain the whole genome sequence of S. aureus by shotgun sequencing (Sanger Institute, UK) which divided into several contigs and by using several bioinformatics tools, the bacterial chromosome sequence can be obtained, aligned and evaluated.
Bioinformatic analysis
The genome sequence of S. aureus was manipulated by using Artemis. This is a free genome browser and annotation tool that allows the visualization of sequence characteristics, next-generation details, and study findings within the sequence (Ali, 2016; Rutherford et al., 2000). Using BIOEDIT sequence alignments, translations, and comparisons were performed. (Hall, 1999). Gene Bank databases search for homologoue sequences and the BLAST algorithm was used. Based on genetic distances, neighbor-joining trees were constructed and calculated by the two-parameter method using MEGA6 (Kimura, 1980; Saitou and Nei, 1987; Tamura et al., 2013). Five hundred bootstrap confidence values estimated the reliability of the trees.
Results
48 S. aureus out of 140 milk samples were isolated from dairy cows and 31 S.aureus out of 100 milk samples were isolated from she- camels. The result of the presence or absence of some resistance genes in S. aureus isolated from cattle (A1, A2, A3, A4, A5, A6, A7, and A10). A1 isolate showed that there were 3 genes, aac(6’)-aph(2’’), mecA, and tet(K) were identically 100 % to its corresponding reference genes, but in the case of tet(38) gene, it was the identically 99.85% (Table 2). The absence of blaZ, aph(3’)-III, ant(6)-Ia, and fusB genes (Table 1). A2 isolate had 3 genes, aac(6’)-aph(2’’) and mecA were identically 100 %, but in the case of tet(38) genes the percent of identically 99.85% compared with reference genes and absences of blaZ, aph(3’)-III, ant(6)-Ia, str, tet(K), and fusB genes (Table 1). A3 isolate, there were 6 genes aph(3’)-III, ant(6)-Ia, mecA, blaZ, fusB and tet(K) were identically 100 % and tet(38) was identically by 99.85% (Table 2) and there were absent of aac(6’), and aph(2’’)genes (Table 1). A4 isolate had 2 genes, mecA and blaZ were identically100 % but, the tet(38) genes identically were 99.85% compared with genes in Gene bank (Table 2), and there were absent of aph(2’’), aph(3’)-III, ant(6)-Ia, tet(K), and fusB genes (Table 1). A5 isolate had 2 genes aac(6’)-aph(2’’) and mecA were identically 100 % but, the tet(38), blaZ and tet(K) were identically 99.85%, 99.23% and 99.89% (Table 2), in addition to, absences of aph(3’)-III, ant(6)-Ia, and fusB genes (Table 1). A6 isolate had 3 genes aac(6’)-aph(2’’) and mecA were identically100 % but in the case of tet(38) and tet(K) genes were 99.85% and 99.89% identical to its reference genes (Table 2), blaZ, aph(3’)-III, ant(6)-Ia, and fusB genes were absent (Table 1). A7 isolate had 3 genes aac(6’)-aph(2’’), mecA and tet(K) were identically100 %, but in case of tet(38) genes was identically 99.85% (Table 2), and there were absent of blaZ, aph(3’)-III, ant(6)-Ia, and fusB genes (Table 1). Strain A10 had only one gene blaZ was identically 100 %, and tet(38) genes was identically 98.97% (Table 2), and there were absence of mecA, aac(6’), aph(2’’), aph(3’)-III, ant(6)-Ia, tet(K),and fusB genes (Table 1). The result of the presence and absence of resistance genes in S. aureus isolated in this study from she-camel included isolates A11, A12, A13, and A15. A11 isolate had only one genes blaZ was identically 100 %, but the tet(38) genes identically 99.97% (Table 2), and the absences of mecA, aac(6’), aph(2’’), aph(3’)-III, ant(6)-Ia, tet(K), and fusB genes (Table 1). A12 andA13 isolates showed that there is one gene tet(38) was identically 99.63 % (Table 2) in addition to the absence of mecA, blaZ, aac(6’), aph(2’’), aph(3’)-III, ant(6)-Ia, tet(K), and fusB genes (Table 1). A15 isolate had one gene blaZ was identically 100 %, but in case of the tet(38) genes was similar 99.97% to its reference gene (Table 2) in addition to the of absence of mecA, aac(6’), aph(2’’), aph(3’)-III, ant(6)-Ia, str, tet(K), and fusB genes (Table 1). Str gene (Aminoglycoside 6-adenylyltransferase) was absent from all isolates of S. aureus originated from cattle and she camels (Table 1).
Evolutionary analysis by Maximum Likelihood method
The evolutionary history was inferred by using the Maxi
Table 1: Presence and absence of resistance genes in different Staphylococci aureus isolate from cattle and camels
| No. | Isolates | Animals | Species |
Presence and absence of resistance genes |
|||||||||
| mecA | blaZ | aac(6') | aph(2'') |
aph(3') -III |
ant(6) -Ia |
str | tet(K) | tet(38) | fusB | ||||
| 1 | A1 | Cattle | S. aureus | 100% | Nil | 100% | 100% | *Nil | Nil | Nil | 100% | 99.85% | Nil |
| 2 | A2 | Cattle | S. aureus | 100% | Nil | 100% | 100% | Nil | Nil | Nil | Nil | 99.85% | Nil |
| 3 | A3 | Cattle | S. aureus | 100% | 100% | Nil | Nil | 100% | 100% | Nil | 100% | 99.85% | 100% |
| 4 | A4 | Cattle | S. aureus | 100% | 100% | Nil | Nil | Nil | Nil | Nil | Nil | 99.85% | Nil |
| 5 | A5 | Cattle | S. aureus | 100% | 100% | 100% | 100% | Nil | Nil | Nil | 99.89% | 99.85% | Nil |
| 6 | A6 | Cattle | S. aureus | 100% | Nil | 100% | 100% | Nil | Nil | Nil | 99.89% | 99.85% | Nil |
| 7 | A7 | Cattle | S. aureus | 100% | Nil | 100% | 100% | Nil | Nil | Nil | 100% | 99.85% | Nil |
| 8 | A10 | Cattle | S. aureus | Nil | 100% | Nil | Nil | Nil | Nil | Nil | Nil | 98.97% | Nil |
|
9 |
A11 | Camel | S. aureus | Nil | 100% | Nil | Nil | Nil | Nil | Nil | Nil | 98.97% | Nil |
| 10 | A12 | Camel | S. aureus | Nil | Nil | Nil | Nil | Nil | Nil | Nil | Nil | 99.63% | Nil |
| 11 | A13 | Camel | S. aureus | Nil | Nil | Nil | Nil | Nil | Nil | Nil | Nil | 99.63% | Nil |
| 12 | A15 | Camel | S. aureus | Nil | 99.88% | Nil | Nil | Nil | Nil | Nil | Nil | 99.56% | Nil |
*Nil=Nothing
Table 2: Identity percent of resistance genes in different Staphylococci aureus isolate
| Isolates | Resistance gene | Identity percent | Query/HSP length | Contig | Position in contig | Predicted phenotype | Accession number |
| A1 | aac(6')-aph(2'') | 100 | 1440 / 1440 | EMBOSS_001 | 2651606..2653045 | Aminoglycoside resistance | |
| mecA | 100 | 2010 / 2010 | EMBOSS_001 | 2756089..2758098 | Beta-lactam resistance | ||
| tet(K) | 100 | 1380 / 1380 | EMBOSS_001 | 2762824..2764203 | Tetracycline resistance | ||
| tet(38) | 99.85 | 1353 / 1353 | EMBOSS_001 | 679533..680885 | Tetracycline resistance | ||
| A2 | aac(6')-aph(2'') | 100 | 1440 / 1440 | EMBOSS_001 | 2774658..2776097 | Aminoglycoside resistance | |
| mecA | 100 | 2010 / 2010 | EMBOSS_001 | 2763972..2765981 | Beta-lactam resistance | ||
| tet(38) | 99.85 | 1353 / 1353 | EMBOSS_001 | 849985..851337 | Tetracycline resistance | ||
| A3 | aph(3')-III | 100 | 795 / 795 | EMBOSS_001 | 2714370..2715164 | Aminoglycoside resistance | |
| ant(6)-Ia | 100 | 909 / 909 | EMBOSS_001 | 2715796..2716704 | Aminoglycoside resistance | ||
| mecA | 100 | 2007 / 2007 | EMBOSS_001 | 2728940..2730946 | Beta-lactam resistance | ||
| blaZ | 100 | 846 / 846 | EMBOSS_001 | 2740917..2741762 | Beta-lactam resistance | ||
| fusB | 100 | 642 / 642 | EMBOSS_001 | 2860095..2860736 | Fusidic acid resistance | ||
| tet(38) | 99.85 | 1353 / 1353 | EMBOSS_001 | 2402116..2403468 | Tetracycline resistance | ||
| tet(K) | 100 | 1380 / 1380 | EMBOSS_001 | 2824205..2825584 | Tetracycline resistance | ||
| A4 | mecA | 100 | 2007 / 2007 | EMBOSS_001 | 1645898..1647904 | Beta-lactam resistance | |
| blaZ | 100 | 846 / 846 | EMBOSS_001 | 2734106..2734951 | Beta-lactam resistance | ||
| tet(38) | 99.85 | 1353 / 1353 | EMBOSS_001 | 2683841..2685193 | Tetracycline resistance | ||
| A5 | aac(6')-aph(2'') | 100 | 1440 / 1440 | EMBOSS_001 | 2086295..2087734 | Aminoglycoside resistance | |
| blaZ | 98.23 | 846 / 846 | EMBOSS_001 | 2647711..2648556 | Beta-lactam resistance | ||
| mecA | 100 | 2010 / 2010 | EMBOSS_001 | 2677031..2679040 | Beta-lactam resistance | ||
| tet(38) | 99.85 | 1353 / 1353 | EMBOSS_001 | 2011056..2012408 | Tetracycline resistance | ||
| tet(K) | 99.89 | 888 / 888 | EMBOSS_001 | 2779341..2780227 | Tetracycline resistance | ||
| A6 | aac(6')-aph(2'') | 100 | 1440 / 1440 | EMBOSS_001 | 2768296..2769735 | Aminoglycoside resistance | |
| mecA | 100 | 2010 / 2010 | EMBOSS_001 | 2719292..2721301 | Beta-lactam resistance | ||
| tet(38) | 99.85 | 1353 / 1353 | EMBOSS_001 | 2089322..2090674 | Tetracycline resistance | ||
| tet(K) | 99.89 | 888 / 888 | EMBOSS_001 | 2761281..2762167 | Tetracycline resistance | ||
| A7 | aac(6')-aph(2'') | 100 | 1440 / 1440 | EMBOSS_001 | 2622102..2623541 | Aminoglycoside resistance | |
| mecA | 100 | 2010 / 2010 | EMBOSS_001 | 2758893..2760902 | Beta-lactam resistance | ||
| tet(38) | 99.85 | 1353 / 1353 | EMBOSS_001 | 1743374..1744726 | Tetracycline resistance | ||
| tet(K) | 100 | 1380 / 1380 | EMBOSS_001 | 2765067..2766446 | Tetracycline resistance | ||
| A10 | blaZ | 100 | 846 / 846 | EMBOSS_001 | 552101..552946 | Beta-lactam resistance | |
| tet(38) | 98.97 | 1353 / 1353 | EMBOSS_001 | 2461719..2463071 | Tetracycline resistance | ||
| A11 | blaZ | 100 | 846 / 846 | EMBOSS_001 | 552101..552946 | Beta-lactam resistance | |
| tet(38) | 98.97 | 1353 / 1353 | EMBOSS_001 | 2461719..2463071 | Tetracycline resistance | ||
| A12 | tet(38) | 99.63 | 1353 / 1353 | EMBOSS_001 | 680449..681801 | Tetracycline resistance | |
| A13 | tet(38) | 99.63 | 1353 / 1353 | EMBOSS_001 | 236737..238089 | Tetracycline resistance | |
| A15 | blaZ | 99.88 | 846 / 846 | EMBOSS_001 | 2775234..2776078 | Beta-lactam resistance | |
| tet(38) | 99.56 | 1353 / 1353 | EMBOSS_001 | 1399661..1401013 | Tetracycline resistance |
mum Likelihood method and Tamura-Nei model (Tamura et al., 2013). The tree with the highest log likelihood (-8731.04) is shown. The initial tree for the heuristic search’s initial tree was obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Tamura-Nei model and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 51 nucleotide sequences. There was a total of 678 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kimura, 1980; Saitou and Nei, 1987; Tamura et al., 2013). All resistant genes sequences were arranged into 2 main clusters. The first clusters contain mecA genes, aminoglycosides, fusB gene, and blaz gene except for A11 and 2 reference genes (AJ302698 & AP003139). The second cluster contains mainly the tet(k) and tet(38) genes which are more similar to each other, except some genes that related to the first cluster such as mecA reference gene (AB512767) and 2 blaz reference genes (AJ302698 & AP003139). The tree revealed that all resistant genes are more or less stable genes such as mecA, aminoglycosides, tet(k), tet(38), and fusB except blaz gene showing more variations (Figure 1).
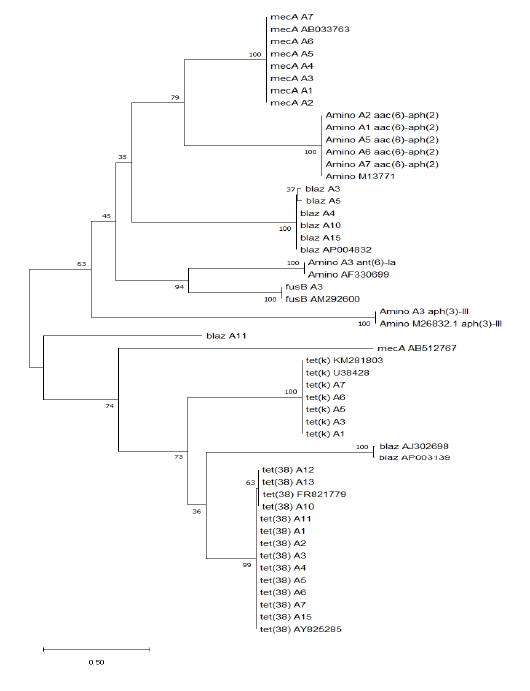
Figure 1: Molecular phylogenetic analysis by maximum likelihood method of resistance genes in different Staphylococci aureus isolates
Discussion
The resistance of many widely used antimicrobial medications is one of the problems facing the management of infection with S. aureus (Bougnom and Piddock, 2017; Luepke et al., 2017). When S. aureus was first discovered, it was simple to treat using available antibiotics. Few years after the use of penicillin in 1940 to treat S. aureus; there were strains of different pathogens resistant to these antibiotics. After that the methicillin was developed and introduced to treat penicillin-resistant S. aureus strains in 1961 (Landecker, 2016). The mode of action of penicillin, and methicillin is very similar and involves inhibiting cell wall synthesis by blocking the peptidoglycan formation. In less than a year after methicillin; strains of S. aureus have been reported to be resistant to methicillin, and those strains have steadily spread globally (Vanderhaeghen et al., 2012; Fair and Tor, 2014).
Conversely, S. aureus strains carrying tetK gene only have been described as resistant to tetracycline, but susceptible to minocycline (Bismuth et al., 1990; Schwarz et al., 1998). The tetM gene is supposed to resist all drugs available in the group, including tetracycline and minocycline. Most isolates with TetM-positive gene and MRSA are typically tetM or tetKM. The tetL gene was only found in isolates already containing tetM gene from S. aureus (Bismuth et al., 1990), however, in vitro S. aureus induces both drug efflux and ribosomal protection (Mojumdar and Khan, 1988; Nesin et al., 1990).
In 1944, aminoglycosides were added, and by the 1950s, strains of S. aureus were aminoglycoside resistant. Through energy-dependent binding to the cell wall and energy-dependent transport through the cytoplasmic membrane, these drugs penetrate bacterial cells, eventually binding to one or more ribosomal sites, thus inhibiting protein synthesis (Maranan et al., 1997). Moreover, it is reported that Staphylococci resistance were resulted from: (1) a chromosomal defect leading to ribosome-binding altered aminoglycosides, (2) inadequate delivery of aminoglycosides into the bacterial cell, causing poor cross-resistance to most aminoglycosides, and, most generally, and (3) aminoglycoside enzyme modulation (Lowy, 2003). In the last case, resistant strains coded for aminoglycoside acetyltransferases, phosphotransferases, and adenyltransferases have the aminoglycoside-modifying genes acc, aph and ant (Maranan et al., 1997; Woodford, 2005). The acetylated, phosphorylated, or adenylated aminoglycosides do not bind to ribosomes and do not inhibit protein synthesis (Woodford, 2005). S. aureus is one of the most common organisms associated with infections among dairy animals with frequent and rapid development of antibiotic resistance. Tetracycline resistance genes have been identified; the main mechanisms conferring resistance to tetracycline to bacteria are active efflux proteins, ribosomal protection proteins, and enzymatic inactivation (Michalova et al., 2004). The aac (6’)-Ie-aph(2’’) gene that inactivates a broad range of the majority of aminoglycosides has also been detected in these isolates in our study. In this study, all resistant genes sequences were arranged into 2 main clusters. The first clusters contain mecA genes, aminoglycosides, fusB gene, and blaz gene except for A11 and 2 reference genes (AJ302698 & AP003139). The second cluster contains mainly the tet(k) and tet(38) genes which are more similar to each other, in addition to mecA reference gene (AB512767) and 2 blaz reference genes (AJ302698 & AP003139). The tree revealed that all resistant genes are more or less stable genes such as mecA, aminoglycosides, tet(k), tet(38) and fusB except blaz gene showing more variations. In Egypt, this work is considered the first record on genetic characterizations based on whole genome sequences of S. aureus isolates, as there was no previous data published on antibiotic resistance. The purpose of this research was to determine the molecular detection of antibiotic genes and the evolutionary phylogenetic relationship of their sequence and clustering.
Conclusion
Techniques of molecular detection continue to be increased to establish antimicrobial resistance trends for S. aureus clinical screening test.
Acknowledgments
We appreciate the financial support by South Valley University, Higher Education, and Scientific Research Sector.
Conflict of interest
The authors declare that there is no conflict of interest for this publication.
Authors contributions
All authors contributed equally to study design, sampling, methodology, interpretation of results, and writing of the manuscript
Ethics statement
All animals were handled by the regulation of the Animal Ethics Committee at the Faculty of Veterinary Medicine, South Valley University, Qena, Egypt, with good animal practice following the guidelines of Research Code of Ethics (RCOE-SVU) at the South Valley University.
References





