Advances in Animal and Veterinary Sciences
Research Article
Detection of Cattle Leptospirosis in Yogyakarta Based on Serology, Molecular, and Histopathological Tests
Tito Suprayoga1, Kurniasih Kurniasih2*, Rini Widayanti3
1Faculty of Veterinary Medicine, University of Gadjah Mada, Jl. Fauna No. 2, Karangmalang, Yogyakarta 55281, Indonesia; 2Departement of Pathology, Faculty of Veterinary Medicine, University of Gadjah Mada, Jl. Fauna No. 2, Karangmalang, Yogyakarta 55281, Indonesia; 3Departemen of Biochemistry, Faculty of Veterinary Medicine, University of Gadjah Mada, Jl. Fauna No. 2, Karangmalang, Yogyakarta 55281, Indonesia.
Abstract | Leptospirosis is a zoonotic disease caused by Leptospira sp. which has been reported around the world. In Indonesia, leptospirosis occurs in humans and animals. Humans can become infected with Leptospira sp. through direct and indirect exposure to the urine of reservoir animals. Rodent (rats) is the main reservoir for Leptospira sp., however, other mammals such as cattle can also be infected with Leptospira sp. and become a source of transmission to humans and other animals. The study aimed to detect pathogenic Leptospira sp. from slaughtered cattle in the abattoir of Yogyakarta, Indonesia. Fifteen sera and kidneys of cattle were fixed in the absolute ethanol. The sera were tested using microscopic agglutination test (MAT), and the kidneys were separated into two part. The first part was extracted, amplified and tested by polymerase chain reaction (PCR) using LipL32 gene primers. The other part was processed by histopathology. Both of these test were used to detect the leptospirosis in cattle. The MAT showed that as much as 33,3% (5/15) of collected samples were seroreactive against Bangkinang serovar. In addition, the PCR showed similar result that indicated by the representation of band product in 498 bp. Histopathology showed that the positive samples in both MAT and PCR test suffering for interstitial nephritis, perivasculitis, atherosclerosis, nephrosis, and fibrosis. This study showed that the MAT, PCR, and histopathology could be used as the detection tools for cattle leptospirosis.
Keywords | Cattle, Histopathology, Leptospira sp., MAT, PCR
Received | October 03, 2020; Accepted | December 03, 2020; Published | January 01, 2021
*Correspondence | Kurniasih Kurniasih, Departement of Pathology, Faculty of Veterinary Medicine, University of Gadjah Mada, Jl. Fauna No. 2, Karangmalang, Yogyakarta 55281, Indonesia; Email: kurniasih_1951@yahoo.co.id
Citation | Suprayoga T, Kurniasih K, Widayanti R (2021). Detection of cattle leptospirosis in Yogyakarta based on serology, molecular, and histopathological tests. Adv. Anim. Vet. Sci. 9(2): 274-279.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.2.274.279
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Suprayoga et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Leptospirosis is a worldwide zoonosis that has a significant impact on animal and public health. During 2018, 285 cases of leptospirosis in humans were reported in Indonesia, with a case fatality rate of 17.8% (Gasem et al., 2020). This zoonotic disease is caused by spirochete bacteria from the genus Leptospira sp. Infection of Leptospira sp. in humans results from direct exposure (with animal tissue, body fluids, or urine) and indirect exposure with soil or urine-contaminated water (Zarantonelli et al., 2018). Leptospira sp. enters the bloodstream through wounds, skin abrasions or it can also be through mucous membranes of the oral cavity and conjunctiva (Brito et al., 2018).
The main reservoir animals for most Leptospira sp. serovars are wild mammals, especially rodents. Domestic animals such as cattle, dogs, sheep, and pigs also act as reservoir animals (Deneke, 2020). Cattle and other ruminants infected with leptospirosis have reproductive problems (Yatbantoong and Chaiyarat, 2019). Cattle and other ruminants infected with leptospirosis have reproductive problems (Sunder et al., 2018), abortion, weak progenies, increased service per conception and calving interval value, weight loss, decreased growth rate, and milk production (Daud et al., 2018). Clinical manifestations of infection with Leptospira sp. depending on the type of serovar infecting and the immune condition of the infected animal (Susanti, 2015).
Leptospira sp. will be located in the kidneys of their natural host such as cattle buffaloe, horse, sheep, goat, pig, dog, and rodent causing little or no damage and maintaining infection in these animals. After infection occurs in the kidney, the cattle will eliminate Leptospira sp. in urine for up to 542 days (Daud et al., 2018; Susanti, 2015).
The prevalence of leptospirosis in beef cattle in Bantul is 18.67%, in Kulon Progo is 14.05% (Susanti, 2015), in the Progo river flow is 13.03% (Mulyani et al., 2016). Serovar Hardjo, Pomona, Icterohaemorrhagiae and, Grippotyphosa are serovars that often infect cattle (Grippi et al., 2020).
Microscopic agglutination test (MAT) is the gold standard for diagnosing leptospirosis recommended by the World Health Organization (WHO) (Fraga et al., 2015). The principle of this test is based on the reaction between the serovar antigen Leptospira sp. with antibodies from serum that can be observed using a dark field microscope to see the presence of agglutination (Levett, 2001). Polymerase chain reaction (PCR) can detect pathogenic Leptospira sp. quickly and precisely. The advantage of using PCR technology is that it has a high test sensitivity. Various sequences have been targeted including the 16S rRNA gene, the LipL32 gene, which encodes the membrane lipoprotein Leptospira sp. and genes coding for the immunoglobulin-like proteins (Lig) Leptospira sp, which are important as virulence factors for these bacteria (Fraga et al., 2015; Marquez et al., 2017). LipL32 was consistently found in pathogenic Leptospira sp. (Podgoršek et al., 2020).
The study aimed to detect pathogenic Leptospira sp. circulating in slaughtered cattle from Yogyakarta abattoir through serological microscopic agglutination tests (MAT), molecular polymerase chain reaction (PCR) and histopathology. It is expected to provide information on the existence of pathogenic Leptospira sp. that infect cattle in Indonesia. Further, this data provide benefits in efforts to prevent the incidence and transmission of leptospirosis in humans and animals.
MATERIALS AND METHODS
Ethic approval
All stages of the research were approved by the Ethical Commite of Gadjah Mada University (number: 0045/EC-FKH/Int./2020). A total of fifteen sera and kidneys from slaughtered cattle were collected from the abattoir in Yogyakarta. The data of collected specimens were embedded in Table 1.
Table 1: The data of collected specimens from the abattoir of Yogyakarta.
| No | Sample code | Origin | Sex | Age | Breed |
| 1 | S1 | Bantul | Male | 2 | Limpo |
| 2 | S2 | Magelang | Male | 1 | Limpo |
| 3 | S3 | Prambanan | Male | 2 | Limpo |
| 4 | S6 | Prambanan | Male | 2,5 | Simpo |
| 5 | S7 | Prambanan | Male | 2 | Limpo |
| 6 | S8 | Sleman | Male | 2,5 | Limpo |
| 7 | S10 | Prambanan | Male | 2,5 | PO |
| 8 | S11 | Prambanan | Male | 3 | Simpo |
| 9 | S12 | Prambanan | Male | 3 | Simpo |
| 10 | S13 | Prambanan | Male | 3 | Simpo |
| 11 | S14 | Prambanan | Male | 2 | Po |
| 12 | S15 | Prambanan | Male | 3 | Simpo |
| 13 | S16 | Prambanan | Male | 3 | Simpo |
| 14 | S17 | Bantul | Female | 5 | Simpo |
| 15 | S18 | Prambanan | Male | 1,5 | Limpo |
Limpo= mixed breed of Limousine cattle × Ongole cattle; Simpo mixed breed of Simmental cattle × Ongole cattle and PO= Ongole cattle.
Time and place of study
The study was conducted from April 2019 until April 2020. The study was conducted in several places. The microscopic agglutination test (MAT) was carried out in the Laboratory of Balai Besar Penelitian dan Pengembangan Vektor dan Reservoir Penyakit (B2P2VRP), Salatiga, Central Java, Indonesia. The PCR was conducted in the Laboratory of Biotechnology, Disease Investigation Centre, Wates, Yogyakarta, Indonesia. The histopathology was conducted in the Department of Pathology, Faculty of Veterinary Medicine, University of Gadjah Mada, Yogyakarta, Indonesia.
Microscopic agglutination test (MAT)
All the sera were tested using microscopic agglutination test (MAT) to detect the presentation of Leprospira sp. antibody (Mulyani et al., 2016). The MAT test used several Leptopira sp. antigens including serovar Bangkinang, Gryppotyphosa, Icterohaemorrhagiae, Canicola, Pyrogenes, Hardjo, Hebdomadis, Pomona, Djasiman, Robinsoni, Bataviae, Mini, Sarmin, Manhao, and Rama. The positive reaction of antigen-antibody was indicated by the titre ≥1:80 of each serovar.
Polymerase chain reaction (PCR) and histopathology
The kidney samples were separated into two parts. The first was tested using polymerase chain reaction (PCR), and the other using histopathology. For the PCR, the kidneys were stored inside the absolute ethanol. Further, the organ was DNA extracted with gSYNCTM DNA Extraction Kit (Geneaid) then amplified. Lipl32 forward primer 5’- GGA CGG TTT AGT CGA TGG AA -3 ‘ and LipL32 reverse primer 5’- GGG AAA AGC AGA CCA ACA GA -3’ were used to amplify LipL32 gene (Ikaratri, 2020). The PCR used 35 cycles with 95oC of first denaturation for 5 minutes, then 94oC of denaturation for 30 seconds, 58oC of annealing for 30 seconds, 72oC of elongation for 1 minute, followed by 72oC of final elongation for 7 minutes, and holding 4oC. On the other hand, the histopathology was performed for the other part of kidney using routine staining (Slaoui and Fiette, 2011).
Analysis data
The data was represented as positive and negative for the MAT and PCR. However, the data of histopathology was reported as the histopatological finding. Further, the collected data were analysed descriptively.
RESULTS AND DISCUSSION
The MAT test showed that 5/15 (33,3%) samples were positive against Leptospira sp. (serovar Bangkinang) (Table 2). The result was categorised as positive if the titre antibody representing between 80. In this study, the titre antibody of the collected specimen between 80 until 320. These results indicated high level of titre antibody. The PCR demonstrated similar results with the MAT (Table 2). The positive samples of PCR was indicated by the appearance of band in of 498 bp according to the target Leptospira sp. (Figure 1). The histopathology showed that the positive sample in MAT and PCR demonstrated interstitial nephritis, glomerulonephritis, perivasculitis, atherosclerosis, nephrosis, and fibrosis (Table 2 and Figure 2). The glossary of MAT, PCR, and histopathology was embedded in Table 2.
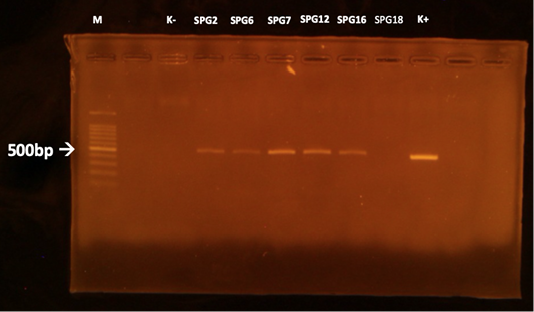
Figure 1: PCR results of the kidneys using LipL32 forward and reverse primers. M: marker; K +: positive control; K-: negative control.
This study revealed that there was one sample (SPG12) that seronegative in MAT but positive in PCR. Next, there was one sample (SPG18) that seropositive in MAT but negative in PCR test. It is suspected by the low sensitivity of MAT in detection of the samples in acute phase of infection (Riyadi and Sunarno, 2019). The MAT test results began to show positive results when samples were collected on days 10-12 after the onset of the disease. In natural infections, it is difficult to detect when the infection started. In SPG18 the infection was detected chronic, as seen from the histopathological changes in the form of fibrosis in the kidney organ, so that the presence of infectious agents in the cattle’s body was not detected using the PCR test. According to Ahmed et al. (2012), most molecular diagnostic tests to detect Leptospira sp infection is based on the amplification of Leptospira-specific nucleic acids from clinical samples that contain Leptospira sp. in the early phase of acute illness.
Table 2: The result of the serological, molecular and histopathology.
| No. | Sample code | Result | ||
| MAT | PCR | Histopathologi | ||
| 1 | SPG 1 | - | - | No histopathological changes |
| 2 | SPG 2 | + | + | Interstitial nephritis |
| 3 | SPG 3 | - | - | No histopathological changes |
| 4 | SPG 6 | + | + | Interstitialis nephritis |
| 5 | SPG 7 | + | + | Interstitial nephritis, glomerulonephritis |
| 6 | SPG 8 | - | - | No histopathological changes |
| 7 | SPG 10 | - | - | No histopathological changes |
| 8 | SPG 11 | - | - | No histopathological changes |
| 9 | SPG 12 | - | + | Interstitial nephritis, perivasculitis, glomerulonephritis |
| 10 | SPG 13 | - | - | No histopathological changes |
| 11 | SPG 14 | - | - | No histopathological changes |
| 12 | SPG 15 | - | - | No histopathological changes |
| 13 | SPG 16 | + | + | Interstitial nephritis, atherosclerosis, nephrosis |
| 14 | SPG 17 | - | - | No histopathological changes |
| 15 | SPG 18 | + | - | Fibrosis |
Leptospira sp infection will be increasing IgM antibodies at the beginning of the infection, followed by IgG antibodies that will last a long time. The MAT test will detect either IgM nor IgG, and this test can be used after 6-10 days of infection. This test is based on the agglutination reaction due to the reaction between Leptospira sp. antigen, and antibody in the patient’s serum, which is observed using a dark field microscope (Grippi et al., 2020). Case reports regarding Bangkinang serovar infection in cattle have never been reported. In 1999 Brenner et al., reported the first isolation of Leptospira interrogans serovar Bangkinang but in cases of human leptospirosis in Indonesia.
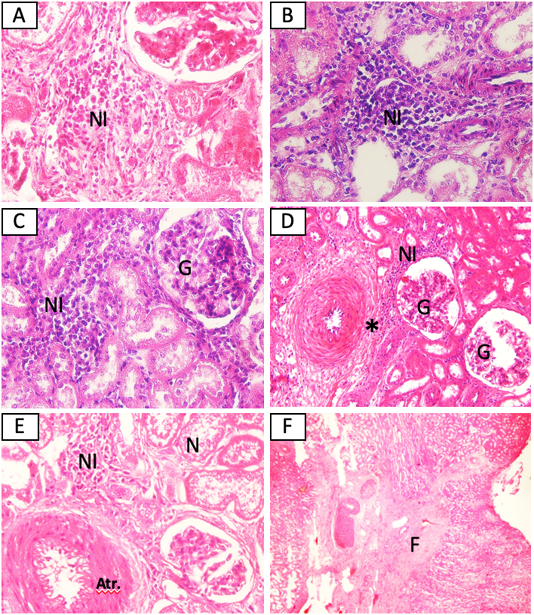
Figure 2: Histopathological changes of kidney from cattle positive with leptospirosis. (A) Sample SPG2, (B) Sample SPG6, (C) Sample SPG7, (D) sample SPG12, (E) Sample SPG16, (F) sample SPG18. NI: Interstitial nephritis, G: glomerulonephritis, *: perivasculitis, N: nephrosis, Atr.: atherosclerosis. H and E (A, B, C, E 40x), (D 20x), (F 4x).
So that the findings of Bangkinang serovar infection in this study are new findings in cases of leptospirosis in cattle. In this case, the cow acts as an incidental host because cattle are maintenance hosts of serovar Hardjo (Grippi et al., 2020). The Bangkinang serovar belongs to the Autumnalis serogroup and Bangkinang I strain (Slack et al., 2005). Serogroup Autumnalis has been detected infecting cattle in North Tunisia, but the serovars that infect are Autumnalis and Bim (Khbou et al., 2016). Autumnalis serogroup is an incidental agent causing leptospirosis in cattle (Martins et al., 2014). Several serovars detected by microscopic agglutination test have infected cattle in the Special Region of Yogyakarta including Icterohaemorrhagie, Canicola, Hardjo, Bataviae and Tarassovi in Bantul; Icterohaemorrhagie, Canicola, Ballum, Cynoptery, Australis, Hardjo, Bataviae, and Tarassovi in Kulon Progo (Susanti, 2015); Hardjo, Rachmawati, Icterohaemorrhagiae, Bataviae, Javanica, Canicola, Pyogenes, Tarassovi, and Celledoni in the Progo River (Mulyani et al., 2016). The detection of leptospirosis in cattle serum from slaughterhouses was also carried out by Sumarningsih et al. (2016) using the microscopic agglutination test. The study has detected Leptospira sp. serovar Hardjo most commonly infects cattle. Other serovars that have been detected are Tarrasovi, Icterohaemorrhagiae, and Bataviae. Serovars that commonly infect cattle are Hardjo, Grippotyphosa, Icterohaemorrhagiae, and Pomona.
The use of PCR can help in the early diagnosis of leptospirosis. Amplification of specific target genes for the detection of leptospirosis has been carried out by previous investigators. Guedes et al. (2019) used primers targeted at the 16S rRNA gene that produced a 330 bp product to identify Leptospira sp. that infects cattle in the Brazilian Amazon. Sumanta et al. (2015) used primers targeted at the 16S rRNA gene to detect leptospirosis in mice in Yogyakarta, Indonesia. Primers targeted at the secY and 16S rRNA genes could identify the Leptospira interrogans serogroup Sejroe serovar Hardjo (Cosate et al., 2017).
The outer protein membrane (OMPs) plays an important role in the pathogenesis process of Leptospira sp. LipL32 is an outer membrane protein and is a specific virulence factor most commonly found in pathogenic Leptospira sp. and not found in non-pathogenic or saprophytic Leptospira sp. (Nagraik et al., 2020). Positive results of PCR testing using primers targeted at the LipL32 gene in this study indicate that Leptospira sp. infecting these cattle is a type of pathogenic Leptospira sp. LipL32 gene can be used as a genetic marker to detect leptospirosis caused by Leptospira interrogans (Pinna et al. 2018). Latifah et al. (2017) stated that the LipL32 gene was found in pathogenic Leptospira sp. The primer design that targets the LipL32 gene can amplify pathogenic Leptospira sp, namely Leptospira interrogans serovar Bataviae, Leptospira interrogans serovar Australis and Leptospira interrogans serovar Javanica by producing a product of 786 bp.
Leptospirosis is characterized by the development of vasculitis, endothelial damage, and inflammatory infiltrates composed of monocytes, plasma cells, and neutrophils. While in the kidney, interstitial nephritis is the major finding accompanied by an intense cellular infiltration composed of neutrophile and monocyte (Yadeta et al., 2016). Leptospirosis induces renal dysfunction such as acute tubulointerstitial nephritis and acute tubular necrosis and also vasculitis but rare (Wu and Wu, 2019).
Pathological reactions in the kidneys due to Leptospira sp. infection are caused by a direct reaction caused by the organism and also due to the body’s immune response to the infection. When Leptospira sp. invasion occurs, bacterial components such as lipopolysaccharides, peptidoglycan, and outer membrane proteins (glycoproteins) will activate the Toll-like receptor-dependent pathway. Toll like receptor activation in the proximal tubular will produce pro inflammatory cytokines and chemokines (include inducible nitric oxide (iNOS), monocyte chemoattractant protein-1 (CCL2/MCP-1), regulated upon activation normal T-cell expressed and secreted (RANTES), and tumor necrosis factor (NTF-α) then trigger the inflammatory process. The cell-mediated immune response through the release of cytokines and chemokines will cause the migration of neutrophils to the glomerulus and interstitial tissue cause glomerulonephritis and tubulointerstitial nephritis. Leptospira sp. will induce the pathway for fibrosis in tubular cells by activating the transforming growth factor-β1/Smad pathway. This activation will result in increased extracellular matrix production in the renal tubular cells (Tanaka et al., 2017; Wu and Wu, 2019).
Histopathological changes in this study were similar to the histopathological findings made by previous investigators. Prakoso et al. (2020) found changes in the kidney organs of cattle infected with Leptospira sp. including chronic interstitial nephritis, hemorrhage, renal vascular congestion, and renal tubular nephrosis. Research conducted by Ajayi et al. (2020) stated that kidneys infected with Leptospira sp. showed changes in the form of tubular necrosis, glomerular, and tubular atrophy of the kidneys, hemorrhage, and interstitial fibrosis. The inflammatory reaction is dominated by the infiltration of lymphoplasmacytic cells. Meanwhile, research conducted by Magalhães et al. (2020) stated that the histopathological changes of the bovine kidney originating from Triangulo Mineiro infected with Leptospira sp. is the occurrence of hyalinize in the renal tubules, congestion, and hydropic degeneration of the renal tubules.
CONCLUSION
Based on serological and molecular tests, it was found Leptospira interrogans serovar Bangkinang at the Yogyakarta slaughterhouse. Further, the positive sample with leptospirosis showed several histopathological changes including interstitial nephritis, glomerulonephritis, perivasculitis, atherosclerosis, tubular nephrosis, and fibrosis in positive bovine kidneys.
Author’s Contribution
Tito Suprayoga was conducted the sample collection, performed the examination and analyzed the data. Kurniasih Kurniasih and Rini Widayanti was performed the molecular and serological tests. All author contributed during writing the draft of manuscript and approved the final version of this manuscript.
Conflict of interest
The authors have declared no conflict of interest.
REFERENCES





