Advances in Animal and Veterinary Sciences
Research Article
Use of Quorum Sensing Gene SdiA as a Molecular Marker for Salmonella Diagnosis
Shaymaa Abdelmalek1, Esraa Abdulmaged Elshafiee2, Wafy Hamed3, Mona Kadry2*
1Department of Microbiology and Immunology, Faculty of Veterinary Medicine, Cairo University, Giza, Egypt; 2Department of Zoonosis, Faculty of Veterinary Medicine, Cairo University, Giza, Egypt; 3Medical Laboratory Specialist, Quesna, Menofiea, Egypt.
Abstract | The alterations in cell density of the bacteria as a result of the gene expression mechanism is called quorum sensing and these changes in Salmonella has been an essential issue for a long time. The quorum sensing gene (sdiA) found in Salmonella spp. is an important regulatory gene for Salmonella survival, colonization and communication with other bacteria and hosts. Salmonella live in the human intestine, which harbors a great density and variety of bacterial cells, in addition to the other flora exist in the colon and all of them communicate amongst themselves and with the host itself to make a change for adaptive processes, such as antibiotic production, invasion of host cells and biofilm formation . Twenty-eight local Egyptian Salmonella isolates from different localities and different sources in Egypt such as, human stool, Egyptian cattle egrets and storks and grilled chicken from electric grills, were tested for the presence of sdiA gene by using PCR and compared with four non-Salmonella local isolates. All Salmonella isolates were PCR-positive for the sdiA gene (274-bp product). All non-Salmonella isolates were PCR-negative for the sdiA gene. Sequencing of sdiA gene revealed thet there were more than 99 % similarity to sdiA gene sequences existing in the GenBank database of different serotypes of Salmonella enterica strains. Therefore, it can be suggested that the SdiA gene is conserved among Salmonella enterica strains regardless of their serotypes. This work provide evidence that the sdiA gene is necessary for Salmonella virulence and the (sdiA) PCR assay is a unique, highly specific molecular marker for the diagnosis and detection of Salmonella.
Keywords | Salmonella, Virulence, Quorum sensing, PCR, Sequencing
Received | September 14, 2020; Accepted | December 03, 2020; Published | December 05, 2020
*Correspondence | Mona Kadry, Department of Zoonoses, Faculty of Veterinary Medicine, Cairo University, Box 12211, Giza, PO, Egypt; Email: mona.kadry.mohamed@cu.edu.eg
Citation | Abdelmalek S, Elshafiee EA, Hamed W, Kadry M (2021). Use of quorum sensing gene SdiA as a molecular marker for Salmonella diagnosis. Adv. Anim. Vet. Sci. 9(1): 15-20.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.1.15.20
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Abdelmalek et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Salmonella spp. are gram-negative bacilli spread among domestic and wild animals and humans that cause severe illnesses, such as diarrhoea, fever and general fatigue (Baired-Parker, 1990). Salmonella spp. are a major food-borne pathogen in animals and humans (Humphery, 2002). Hence, rapid detection of Salmonella in clinical samples, foodstuffs and water is critical for prompt and accurate diagnosis of Salmonella, which in turn helps in control and prevention of the disease. Molecular diagnosis by using PCR assay is more accurate and reliable technique for the detection of pathogens. Interestingly, several PCR techniques have been developed for detection of numerous genes in Salmonella, for example (Rahn et al., 1992), agfA (Doran et al., 1993), hilA and sirA (Guo et al., 2000) 16S rRNA (Iida et al., 1993), viaB (Hashimoto et al., 1995), and ttr (Malorny et al., 2004). However, the invA gene is the most commonly used for the diagnosis of Salmonella due to it has unique DNA sequences (Hara-Kudo et al., 2005; Abdel-Aziz, 2016). The majority of Salmonella spp., except S. litchfield and S. Senftenberg, are found to harbour invA gene. In the invasion-associated protein secretion mechanism, the invA gene encodes an essential compound (Gala’n et al., 1992). Besides, the invA gene has been found to be located in region that constitutes a pathogenicity island –PAI-, and it is in some Salmonella serotypes unstable, such as S. litchfield and S. Senftenberg (Ginocchio et al., 1997).
Although, in the past, many researches have revealed that most of the bacteria use several quorum-sensing systems, but the biological significance of this phenomenon remains unknown. Bacterial cells are able to modify the gene expression pathway in response to extracellular signals. In contrast, certain bacteria that can release and detect signalling compounds to control gene expression using quorum sensing were first detected in marine Vibrio fischeri (Nealson and Hasting, 1979). Quorum sensing is the communication phenomenon between bacteria, and it controls cellular aggregation-dependent factors, such as antibiotic production, invasion of host cells and biofilm formation (Wei and Zhao, 2018). The bacteria secrete acyl homoserine lactone (AHL), which is a species-specific autoinducer-1 (AI-1). AI-1 is an extracellular signalling molecule that is responsible for the activation of quorum sensing (Zhang and Li, 2016). Gene transcription, which is regulated by the regulatory gene sdiA (LuxR family), is induced as soon as the threshold level is reached.. The sdiA gene of Salmonella (LuxR family) regulates the intestinal survival, colonization and invasion (pathogenicity) of Salmonella (Ahmer, 2004). Host entrance and adherence to intestinal mucosa are important for Salmonella pathogenicity in cell invasion. There are interacting signals between Salmonella and the host. These signals lead to changes in ruffling of the membrane and the entry of Salmonella (Gala’n, 1995). Therefore, the development of a molecular marker for Salmonella spp. detection is very important.
The aim of the current study was to examine the possibility of using the sdiA gene as a molecular marker for diagnosis of local Egyptian Salmonella strains recovered from different sources, including human stool, foodstuffs, and carriers gathered from the Cairo, Giza and Menofeia governorates. Moreover, sequencing of the sdiA gene was used to detect the epidemiological link between the different Egyptian Salmonella isolates.
MATERIAL AND METHODS
Bacterial isolates
Twenty-eight local Egyptian Salmonella isolates from the Cairo, Giza and Menoufia governorates were collected from different sources, such as 75 human stool from clinically affected persons tested by the Widal test, 35 Egyptian stork (major Salmonella carriers) and 30 from multilayer electric grills, grilled chicken. All strains of Salmonella were characterized culturally, biochemically, serologically and molecularly by using a PCR assay for the invA gene as previously studied by (Abdelmalek et al., 2019), as it is the most common PCR assay for Salmonella isolate identification,. Four Local non-Salmonella bacterial isolates were used as negative controls for the sdiA gene as follows: E. coli, Klebsiella pneumoniae, Proteus vulgaris and Pseudomonas aeruginosa. They were local isolates and were identified culturally, biochemically and molecularly.
The extraction of DNA
On nutrient agar medium (SIGMA), the local Salmonella isolates were grown, and the bacterial cells were harvested from the culture plates using a sterile cotton swab placed in an Eppendorf tube containing 1X PBSS. [1X phosphate buffered saline prepared by dissolving one tablet in 100 ml of distilled water (SIGMA)], and mixed well. The specimens were then centrifuged for 5 min at 9000 x g (washing was repeated three times with PBS) and then washed once with sterile distilled deionized water. Then, at 9000 x g for 5 min, centrifugation was performed and the supernatant was extracted. Subsequently, 100 μl of sterile deionized distilled water was suspended in the bacterial pellet. The samples were put in a heat block (Grant-Bio, England) for 5 min at 100OC and then cooled in ice water immediately. The specimen had been centrifuged, (Wang et al., 1996).
PCR amplification and electrophoresis
In a 25 μl reaction mixture containing 12.5- μl of 2X master mix - (EmeraldAmpGT, A2201-1, TAKARA, Japan), 0.5 μl (25 pmol) of each primer (sdiA F and R), 2 - μl of the extracted DNA and 9.5 - μl of free water from nuclease (Thermo Science, # R0581, USA), PCR amplification was performed.
The cyclic program consisted of denaturation for 5 min at 94oC, followed by 30 cycles of 94oC for 30 s, 52oC for 40 s and 72oC for 0.5 min. Then, final extension at 72oC for 7 min was carried out (Halasti et al., 2006). In electrophoresis using 1.5 percent (w/v) agarose gels (SERVA, Russia), all reaction products in 1X TAE running buffer (SERVA, Russia) were analyzed at 100 V for 30 minuts and imaged using a Spectroline UV transilluminator (Slimline Series, USA).
DNA sequencing
The amplified fragments of sdiA gene in the five selected Salmonella isolates that randomly selected from grilled chicken, human and stork in the same locality were sent to be sequenced at the Animal Health Research Institute (AHRI) in Dokki- El Giza . Using the GeneJET PCR Purification Kit ( Thermo Scientific), the PCR products were purified and a DNA sequencer was used to make the sequencing. The Large Dye Terminator (V3.1 Cycle) Sequencing Kit was used to perform the sequencing step, (Applied Biosystems). The gene sequences have been submitted in the National Center for Biotechnology Information (NCBI) GenBank database by using BankIT tool under the accession numbers MF942870, MF942871, MF942872, MF942873, MF942874 for the human stool, stork and grilled chicken-derived sequences, respectively.
The sequences obtained were determined by a BLAST analysis with GenBank (NCBI). To determine the relationship of our gene sequences recovered from various sources, phylogenetic tree analysis was used and related sdiA gene sequences obtained from the GenBank database. For Clustral W- multiple alignment, BioEdit software was used, and MEGA7 software was used using the maximum likelihood approach for the construction of phylogeny.
RESULTS
PCR results
All local Egyptian Salmonella isolates were PCR-positive for the sdiA quorum sensing gene (274-bp PCR product). The local non-Salmonella isolates E. coli, K. pneumoniae, P. vulgaris and P. aeruginosa were PCR-negative for the sdiA gene (Figure 1).
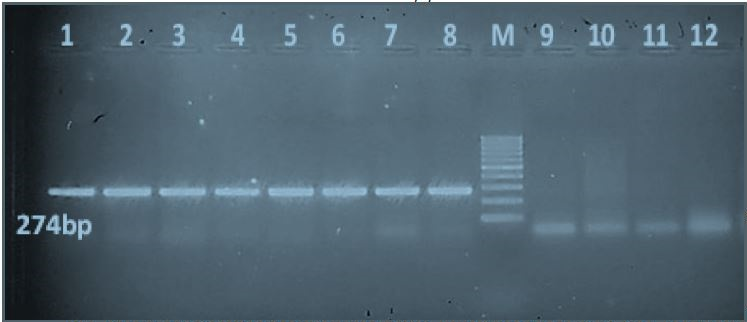
Figure 1: PCR results of the quorum sensing sdiA gene of local Egyptian Salmonella isolates in comparison with non-Salmonella isolates: Lanes 1-8: Salmonella isolates that were PCR-positive for the sdiA gene, which produces a 274-bp product. Lane M: 100-bp molecular ladder. Lanes 9-12: Negative PCR results for the sdiA gene of four non-Salmonella isolates (E. coli, Klebsiella pneumonia, Proteus vulgaris, and Pseudomonas aeruginosa).
Table 1: Oligonucleotide sequence of the quorum sensing sdiA gene.
| Gene | Sequence | Product | Ref. |
|
sdiA F: |
5-AAT ATC GCT TCG TAC CAC-3 | 274 bp |
(Halatsi et al., 2006) |
|
sdiA R: |
5-GTA GGT AAA CGA GGA GCA G-3 |
Sequencing results
In this analysis, the representative sequences were deposited under accession numbers MF942870, MF942871, MF942872, MF942873, and MF942874 for the human stool, stork and grilled chicken-derived sequences, respectively into the GenBank database. The sdiA gene nucleotide sequence data available in GenBank were selected based on the criteria of the available information regarding the source, location and year of isolation (Table 2). The five amplified sdiA gene PCR products were sequenced and matched with the other associated GenBank NCBI-BLAST gene sequences of sdiA.The ≤ 99% similarity to the sdiA gene of Salmonella enterica strains showed that the primers included here exclusively amplifies the 274 bp fragment of the target gene. The phylogenetic analysis results of 5 Salmonella enterica isolates based on sdiA gene sequences are shown in Figure 2. Initially, the sdiA gene sequences of these isolates were compared to those published in GenBank for Salmonella enterica strains (Table 2).
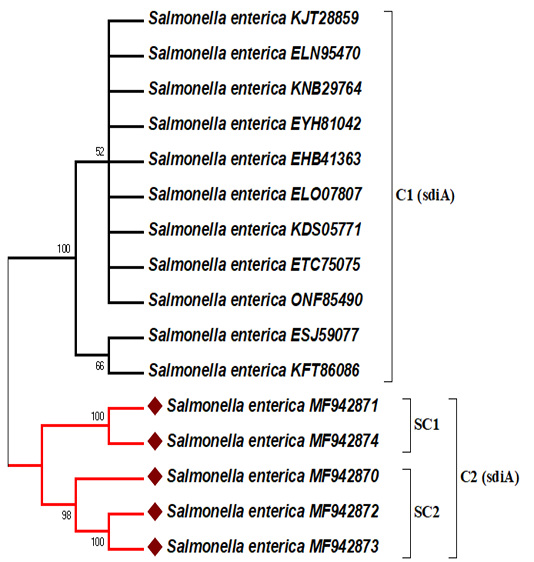
Figure 2: Maximum likelihood phylogenetic tree based on sdiA gene sequences showing the relationship between our study sequences and 11 representatives of sequences retrieved from GenBank. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. Only branches with bootstraps greater than 50 are labelled. The bacterial isolate sequenced in this study is indicated by a red diamond.
DISCUSSION
Salmonella is a very significant food-borne pathogen that causes many public health problems. Prompt and accurate diagnosis is crucial for the detection of Salmonella spp. Molecular diagnosis is more reliable and accurate for pathogen diagnosis than other methods.
Table 2: Isolation source, location and accession no. of Salmonella spp.
| Sample no. | Serotype | Isolation source | Location | Accession no. |
| 1 |
Salmonella Heidelberg |
Environmental sample | USA | KJT28859 |
| 2 |
Salmonella Enteritidis |
Human | USA | ELN95470 |
| 3 |
Salmonella Infantis |
Food sample | Portugal | KNB29764 |
| 4 |
Salmonella Heidelberg |
Chicken breast | USA | EYH81042 |
| 5 |
Salmonella Infantis |
Human | USA | EHB41363 |
| 6 |
Salmonella Enteritidis |
Chicken breast | USA | ELO07807 |
| 7 |
Salmonella Heidelberg |
Human | USA | KDS05771 |
| 8 | Salmonella enteric | Bostourus (faecal sample) | USA | ETC75075 |
| 9 |
Salmonella Typhimurium |
Chicken meat | Brazil | ONF85490 |
| 10 | Salmonella enteric | Dry milk | London | ESJ59077 |
| 11 |
Salmonella Bareilly |
Food sample | USA | KFT86086 |
| 12* |
Salmonella Typhimurium |
Human stool | Egypt | MF942870 |
| 13* |
Salmonella Typhimurium |
Stork (carrier) | Egypt | MF942871 |
| 14* |
Salmonella Typhimurium |
Grilled chicken | Egypt | MF942872 |
| 15* |
Salmonella Typhimurium |
Grilled chicken | Egypt | MF942873 |
| 16* |
Salmonella Typhimurium |
Grilled chicken | Egypt | MF942874 |
*Gene sequences enrolled in our study.
Many Salmonella studies have been focused on the invA gene in different sources; including, animals and humans from different serotypes (Herbert and Hensel, 2004).
Recently, quorum sensing is an essential regulator of bacterial pathogenicity, particularly in Salmonella (Gala’n, 1995; Halasti et al., 2006). The detailed mechanism is still unknown. The sdiA gene is a type of Salmonella regulatory receptor (LuxR family) that is present in the centrosome 42 region (far from the centrosome 63 region of the invA gene) and it does not regulate the virulence-associated secretion mechanism but does regulate essential factors that control intestinal survival and colonization. Until now, the sdiA gene has been shown to initiate genes that help only one of the Salmonella bacteria adhere to the intestinal mucosa (Ahmer, 2004). In the present study, the sdiA gene was examined as a molecular marker for diagnosis of local Egyptian Salmonella isolates from different sources, and compared with non-Salmonella local isolates E. coli, K. pneumoniae, P. vulgaris and P. aeruginosa (Figure 1). Based on the aforementioned data, this study proved that the sdiA gene is highly specific for Salmonella spp. and could be used as a molecular marker for Salmonella diagnosis (Halasti et al., 2006).
DNA sequencing was accomplished to confirm that sdiA gene is a conserved gene in salmonella spp., and predict the epidemiological relationships between our study sequences and related sdiA gene sequences from the available GenBank NCBI-BLAST database. Among the study sequences, there was more than 99 percent matches to sdiA gene sequences of different serotypes of Salmonella enterica strains existing in the GenBank database. These results suggest that the SdiA gene is conserved among Salmonella enterica strains regardless of their serotypes. Previous studies supported this suggestion by sequencing and analysing phylogenetic trees based on the sdiA gene (Herbert and Hensel, 2004; Campos-Galvão et al., 2015). Phylogenetic analysis of the amino acid sequences revealed the existence of two major clusters (C1 and C2) according to the geographical isolation with high bootstrap support value. These geographical clusters may be due to the sdiA gene partial sequence data and are scarce in Africa and the Middle East. Therefore, our study sequence isolates from Egypt with accession numbers MF942870, MF942871, MF942872, MF942873, and MF942874 are located on the same cluster (C2) and show no homology with other sdiA-related sequences (Figure 2). The majority of the sdiA gene data available in GenBank are from the USA and Latin America (Table 2). Egyptian isolates with the same serotype (Salmonella typhimurium) were divided into 2 subclusters (SC1 and SC2) with high bootstrap support values. SC1 shows that the study sequences of stork (wild bird) (MF942871) and the grilled chicken (MF942874) are highly similar to each other, which reflects the potential role of wild birds in the indirect transmission of Salmonella to humans through foodstuffs. This scenario spotlights on the public health implication of the distribution of wild birds, for example, storks, near restaurants in Egypt. Epidemiologically, wild birds that are attracted to human sewage outfalls may disseminate Salmonella to susceptible individuals through faecal shedding and shared environments and via direct contact (Afema and Sischo, 2016; Ahmed et al., 2019). In addition, we cannot deny the role of infected humans in the contamination of ready to eat food, and similar strains of Salmonella typhimurium were found in human stool (MF942870) and grilled chicken (MF942872, MF942873), as shown in SC2 (Figure 2). This finding confirmed that cooks or food handlers who do not follow proper personal hygiene procedures may act as potential sources of recontamination of food after cooking.
Table 3: the different serotypes of the salmonella isolates.
| Isolates/ numbers | Serotypes | No. (%) |
| Human/ 7 |
S. Typhimurium |
3 (42.8) |
|
S. Enteritidis |
1 (14.3) | |
|
S. Virchow |
1 (14.3) | |
|
S. Haifa |
1 (14.3) | |
|
S. Kentucky |
1 (14.3) | |
| carriers stroke/ 10 |
S. Typhimurium |
10 (100) |
| grilled chicken/ 11 |
S. Typhimurium |
10 (100) |
ACKNOWLEDGEMENTS
The authors are thankful to the Department of Zoonoses and the Department of Microbiololgy, Faculty of Veterinary Medicine, Cairo University, Egypt for providing necessary facilities for this study. The authors did not receive any fund for this study.
Author’s Contribution
Shaymaa Abdelmalek, Esraa Abdulmaged.Elshafiee,Wafy Hamed, Mona Kadry contributed to the collection of samples, isolation of strains, performing the molecular detection of target genes, analysis and interpretation of the data as well as writing the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST
The authors have declared no conflict of interest.
REFERENCES





