Advances in Animal and Veterinary Sciences
Research Article
Molecular Differential Diagnosis between Opisthorchis felineus and Metorchis bilis
Aiganym Bekenova1, Ainura Smagulova1, Aleksey Katokhin2, Sergei Borovikov1, Vladimir Kiyan1*
1Research Platform of Agricultural Biotechnology, S. Seifullin Kazakh Agrotechnical University, Nur-Sultan, Kazakhstan; 2Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia.
Abstract | Two species of opisthorchiid trematodes, Opisthorchis felineus (Rivolta, 1884) and Metorchis bilis (Braun, 1890), are known to infect humans and piscivorous mammals in Kazakhstan, causing opisthorchiasis or metorchiasis, respectively. There is difficulty in their differential diagnosis since their eggs are morphologically undistinguished and serological assays are low sensitive and specific. DNA sequenceing, especially for the nuclear ribosomal internal transcribed spacer 2 gene (ITS2), has already been used to differentiate between trematodes of the family Opisthorchiidae. The aim of this study is to establish a method for differentiation and identification of opisthorchiid species using molecular approach. Specific primers for a multiplex PCR were designed based on corresponding ITS2 gene sequence, and its detection limit was assessed by serial dilutions of the genomic DNAs of trematodes. Furthermore, samples of various parasites species of carnivores were tested using the developed assay. In this study, we developed species-specific primers for the differential detection of the two medically important trematodes based on the PCR assay. The species-specific primers were targeted to either variable or conserved regions of the ITS2 gene. ITS2 fragments with different sizes were amplified; 207 bp for O. felineus and 167 bp for M. bilis. Specificity was tested and no cross-reaction occurred and the sensitivity ranged between 2 and 1 ng of DNA template. The developed species-specific primers and PCR assay in this study could be applied for differential diagnosis of these medically and veterinary important opisthorchiid infections.
Keywords | Opisthorchis felineus, Metorchis bilis, Molecular identification, ITS2
Received | August 01, 2020; Accepted | August 7, 2020; Published | August 28, 2020
*Correspondence | Vladimir Kiyan, Research Platform of Agricultural Biotechnology, S. Seifullin Kazakh Agrotechnical University, Nur-Sultan, Kazakhstan; Email: vskiyan@gmail.com
Citation | Bekenova A, Smagulova A, Katokhin A, Borovikov S, Kiyan V (2020). Molecular differential diagnosis between Opisthorchis felineus and Metorchis bilis. Adv. Anim. Vet. Sci. 8(s3): 27-32.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.s3.27.32
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 Bekenova et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Liver flukes of the family Opisthorchiidae are causing serious disease worldwide, with severe complications such as cholangiocarcinoma, choledocholithiasis and pancreatitis (Armignacco et al., 2013; MacLean et al., 1996; Sripa et al., 2011; IARC, 1994). All species within the family are obligate endoparasites with a complex 3-host life cycle including Bithyniidae snails and Cyprinidae fishes as the first and the second intermediate hosts and various piscivorous mammals and birds as definitive hosts including humans (King and Scholz, 2001; Kaewkes, 2003; Fedorova et al., 2016; Tang et al., 2016). Infection has been reported in humans and/or animals within 13 countries of the European Union (Pozio et al., 2013). The disease is endemic and represents a public health problem worldwide including Russian Federation, Ukraine, Belarus and Kazakhstan (Control of Food Borne Trematode Infections, 1995). Prevalence of opisthorchiasis in human reached a peak 2,521 cases in 2002, and gradually decreased to 1,225 cases in 2011 in Kazakhstan (Sultanov et al., 2014).
In veterinary medicine, opisthorchiasis infection was often underestimated in spite of the available reports indicate the widespread prevalence of this infection in wild (fox, corsac) and domestic animals (cats and dogs) in Kazakhstan as consequence of infection Cyprinidae fishes (Sultanov et al., 2014; Beer, 2005; Kiyan et al., 2018). Previous studies in an endemic region revealed that 72% of village dogs and 33% of cats infected with either O. felineus or M. bilis. While, studies on locally caught fish revealed that 9% roach (Rutilus rutilus), 72% ide (Leuciscus idus) and 2.5% bream (Abramis brama) infested with trematode metacercariae (Sultanov et al., 2014).
Methods for diagnosing of the opisthorchiasis disease have some drawbacks as faecal examination for detection of parasite eggs and serological studies by ELISA are very laborious and failed to accurately distinguish between pathogen species (Teimoori et al., 2017; Waikagul et al., 2002; Nöckler et al., 2003). Therefore, the aim of this study is to establish a method for the differentiation and identification of opisthorchiid species using molecular approach; PCR assay and DNA sequencing.
MATERIALS AND METHODS
Samples collection and identification
The adult worms of O. felineus and M. bilis were collected from the liver and gall bladder of artificially infected Syrian hamsters using metacercariae isolated from ides (Leuciscus idus) caught in the Akmola region of Kazakhstan. Additionally, in the current study were used the adult worms of O. felineus and M. bilis collected from the liver of infected foxes from Karaganda region, Kazakhstan. The recovered adult worms were identified under a light microscope based on the morphological properties (Kiyan et al., 2018). After that, adult worms were intensively washed in 1× PBS before being preserved in 70% alcohol and kept at −20°C for DNA extraction. Metacercaria of Holostephanus dubinini, Metorchis xanthosomus and Posthodiplostomum cuticola trematodes pathogens isolated from ides were also used as negative controls. DNA was extracted from one adult worm of Trichinella spiralis, Toxocara canis and Taenia hydatigena in order to examine whether DNA of various parasites species of carnivores of good quality and quantity to be used for PCR. The biosafety ethics for this research was approved by the Animal Ethics Committee of Veterinary Medicine Faculty of KATU (Ethical approval letter, No. 1, 09.11.2017).
DNA extraction and PCR assay
After morphology identification, DNA was extracted from each individual adult worm using the Bio Silica DNA extraction kit (Novosibirsk, Russia), according to manufacturer’s instructions. The quantity and purity of the extracted DNA was determined by measuring absorption at 260 nm and 280 nm in the Nano Drop 2000 (Thermo scientific, USA). DNA was dissolved in ddH2O and stored at – 70°C till use. The ITS2 gene of O. felineus and M. bilis species was amplified using sets of primers, forward (5′-GAACATCGACATCTTGAACG-3′) and reverse (5′-GGAACGACCTGAACACCA-3′) (Kang et al., 2008; Katokhin et al., 2008). Amplification of the marker gene was carried out in a final reaction volume of 50 μl containing 1× Phusion HF buffer, 2.5 mM MgCl2, 1 U Phusion DNA polymerase and 200 μM dNTPs (New England BioLabs Inc., Ipswich, Massachusetts, United States), 25 pmol of each primer and 20 ng of extracted trematode DNA from a single liver fluke specimen as template. The negative control for all experiments was the multiplex PCR reaction with no genomic DNA. PCR was performed as previously described (Kiyan et al., 2018). The resulting PCR products were separated by electrophoresis on an ethidium bromide containing 1.5% agarose gel using 1× TAE buffer solution.
Sequencing and primer design
After amplification of the genome DNA from O. felineus and M. bilis with the ITS2 primers, the resulting 525 bp products were subjected for sequencing and sequence analysis. The PCR-amplified target gene fragment was purified using a E.Z.N.A. Gel Extraction kit (Omega Biotek, Norcross, Georgia, USA), following the manufacturer’s protocols. Sequencing was carried out using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applide Biosystems). The obtained nucleotide sequences were visually checked by the Bio Capt program version 11.0. Subsequently, specific primers for the amplification of the variable region of the ITS2 gene of the two most frequent indigenous opisthorchiid trematode species (O. felineus and M. bilis) were constructed by means of the free available primer programs Perl Primer v1.1.21 (http://perlprimer.sourceforge.net) and Oligo Analyzer 1.2 (http://www.genelink.com). To specifically detect the DNA of O. felineus, sets of primers were selected: OF-spec-F (5′- TGGCATGATTTCCCCACGCA-3′) and OF-spec-R (5′-GCATTGCCAACACTGGAGCC-3′). The specific amplification of M. bilis DNA was performed with the primers MB-spec-F (5′-TGGCATGATTTCCCCGCACG-3′) and MB-spec-R (5′-CGCGCACATACACAACAATAAG -3′).
Sensitivity and specificity test
Amplification of partial ITS2 gene was carried out in a final reaction volume of 25 μl containing 1× Hot Start PCR Buffer (20 mM KCl, 5 mM (NH4)2SO4, 20 mM Tris-HCl, pH 8.3), 2.5 mM MgCl2 and 1 U Maxima Hot Start Taq polymerase (Thermo scientific, USA), 200 mM of dNTP, 100 pmol of each ITS2 primer and 30 ng extracted trematode DNA from a single opisthorchiid liver fluke specimen as template. DNA prepared from trematode samples isolated from ide, Golden hamster and red fox were subjected to the species-specific PCR as control templates. PCR cycling was performed as follows: denaturation at 95°C for 5 min, followed by 35 cycles with 30 s denaturation at 95 °C, primer annealing at 59°C for 40 s, and extension at 72°C for 50 s with a final extension during 5 min at 72°C.
The amplification of the conserved region of the partial ITS2 gene of O. felineus and M. bilis using 100 pmol of each specific primer pair (OF-spec-F/R and MB-spec-F/R primers) was carried out in a 25 μl reaction volume as described above. To examine the specificity, the DNA was prepared from three specimens of both O. felineus and M. bilis, which had been isolated from various definitive hosts. In addition, DNA from O. felineus was tested with MB-spec-F/R primers and DNA of M. bilis with OF-spec-F/R primers in order to rule out the cross-reactivity between primers and non-specific DNA. The primers were also tested with host DNA of the ide, different metacercariae species found in fish, and with the DNA from the red fox and different helminth species of carnivores. The sensitivity of the OF-spec-F/R and MB-spec-F/R primers was checked by subsequent dilution of the DNA of both O. felineus and M. bilis in a 25 μl reaction volume as described above: 20 ng, 10 ng, 8 ng, 6 ng, 4 ng, 2 ng and 1 ng. Each dilution was tested with the respective specific primer pair.
RESULTS
Sequencing and primer design
The amplification of the genomic DNA from O. felineus and M. bilis revealed products of approximately 525 bp using ITS2 primers (Figure 1). The PCR products of opisthorchiid liver fluke species were subjected to sequence analysis. The nucleotide sequences of the studied species were deposited in NCBI GenBank data base under the accession numbers (O. felineus isolate MG952281 and M. bilis isolate MG952282).
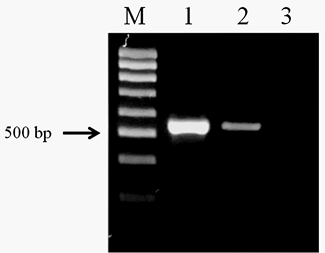
Figure 1: Electrophoretic analysis of PCR products obtained with DNA of the adult trematodes: Lane M, DNA ladder (bp); lane 1, O. felineus DNA; lane 2, M. bilis DNA; lane 3, negative control.
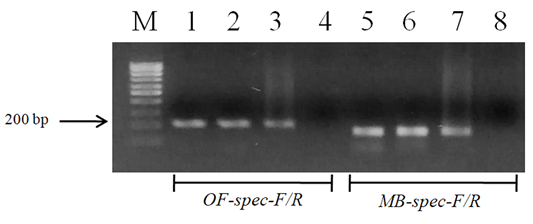
We successfully designed two pair of species-specific primers to amplify the ITS2 fragment of either O. felineus (207 bp) or M. bilis (167 bp) in PCR analysis as expected (Figure 2). The O. felineus ITS2 fragment corresponded to region between positions 71 and 274 and that of M. bilis between positions 71 and 290.
Sensitivity and specificity PCR assay
The designed primers sensitivity was confirmed using three different specimens of O. felineus and M. bilis which had been isolated from various definitive hosts (Table 1). The results are shown in Figure 3 that revealed that no cross-reactivity when testing the MB-spec-F/R primers with O. felineus DNA and the OF-spec-F/R primers with M. bilis DNA (Figure 4).
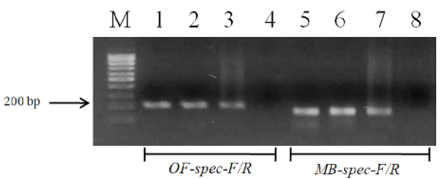
Figure 3: Electrophoretic analysis of PCR products of O. felineus and M. bilis specimens from different definitive hosts: Lane M, DNA ladder (bp); lane 1, O. felineus (ides); lane 2, O. felineus (golden hamster); lane 3, O. felineus (red fox); lane 5, M. bilis (ides); lane 6, M. bilis (golden hamster); lane 7, M. bilis (red fox); lane 4, 8, negative control.
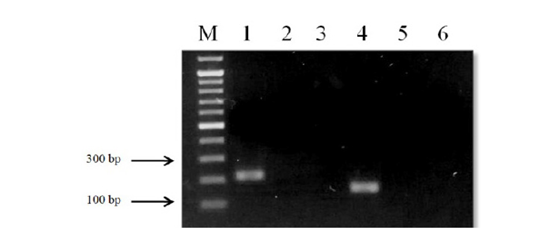
Figure 4: Results of cross-reactivity test of liver flukes DNA and species-specific primers: Lane M, DNA ladder (bp); lane 1, O. felineus DNA and OF-spec-F/R primers; lane 2, O. felineus DNA and MB-spec-F/R primers; lane 4, M. bilis DNA and MB-spec-F/R primers; lane 5, M. bilis DNA and OF-spec-F/R primers; lane 3, 6, negative control.
Table 1: Species used and origin of samples analyzed.
| Species | Host | Origin and region |
| Opisthorchis felineus |
Ides (Leuciscus idus) |
Metacercariae; Akmola region, Kazakhstan |
|
Golden hamster (Mesocricetus auratus) |
Experimental infection; Akmola region, Kazakhstan | |
|
Red fox (Vulpes vulpes) |
Karaganda region, Kazakhstan | |
| Metorchis bilis |
Ides (Leuciscus idus) |
Metacercariae; Akmola region, Kazakhstan |
|
Golden hamster (Mesocricetus auratus) |
Experimental infection; Akmola region, Kazakhstan | |
|
Red fox (Vulpes vulpes) |
Karaganda region, Kazakhstan |
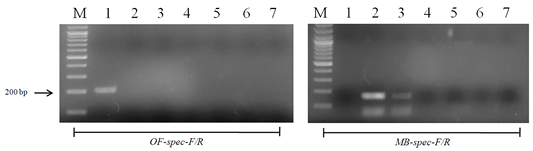
Figure 5: Results of cross-reactivity test of the intermediate host and different metacercariae DNA and species-specific primers: Lane M, DNA ladder (bp); lane 1, O. felineus; lane 2, M. bilis; lane 3, M. xanthosomus; lane 4, H. dubinini; lane 5, P. cuticola; lane 6, L. idus; lane 7, negative control.
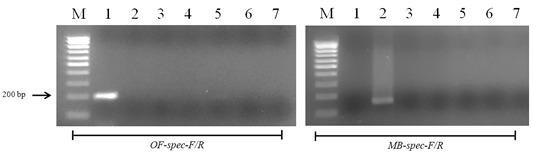
Figure 6: Results of cross-reactivity test of the intermediate host and different helminthes DNA and species-specific primers: Lane M, DNA ladder (bp); lane 1, O. felineus; lane 2, M. bilis; lane 3, T. spiralis; lane 4, T. canis; lane 5, T. hydatigena; lane 6, red fox DNA; lane 7, negative control.
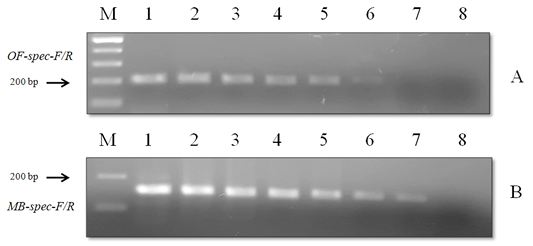
Figure 7: Results of the sensitivity test of species-specific primers with (A) O. felineus DNA and (B) M. bilis DNA: Lane M, DNA ladder (bp); lane 1, 20 ng; lane 2, 10 ng; lane 3, 8 ng; lane 4, 6 ng; lane 5, 4 ng; lane 6, 2 ng; lane 7, 1 ng; lane 8, negative control.
When using DNA of the ide (intermediate host), positive results were obtained only with metacercariae of opisthorchiid (Figure 5). A similar result was obtained when using the DNA of red fox (definitive host) and Trichinella spiralis, Toxocara canis and Taenia hydatigena (Figure 6). The sensitivity of the OF-spec-F/R primers with DNA from O. felineus was detected after dilution down to an amount of 2 ng. M. bilis DNA could be detected down to a level of 1 ng when using MB-spec-F/R primers (Figure 7).
DISCUSSION
The current work is the first in our country to develop the ITS2-targeted PCR-based species-specific diagnosis for two medically important opisthorchiid trematodes - O. felineus and M. bilis. Although there are two other medically important trematodes of the family Opisthorchiidae - O. viverrini and Clonorchis sinensis (both are causing agent of opisthorchiasis-like diseases endemic in Southeast Asia) they were not included in our study because they do not generate natural foci in Kazakhstan (Beer, 2005; Kaewkes, 2003). The main medical and veterinary task is to detect the causing agents of opisthorchiasis and related diseases naturally occurring in Kazakhstan, namely two opisthorchiids: O. felineus and/or M. bilis. Based on eggs morphology, coproovoscopic approach often do not allow to reliably distinguish of pathogens, especially in case of co-infections in human and animals. While, with the use of PCR techniques employing ITS2 sequence differences and correspondingly designed species-specific primers, we can suggest a novel molecular tool to differentiate these pathogenic species.
The species-specific primers produce fragments of approximately 207 and 167 bp in case of adult O. felineus and M. bilis, respectively, regardless the origin of worms from different definitive hosts. The O. felineus-specific and M. bilis-specific amplicons can be easily differentiated by size using 1.5-2% agarose gel electrophoresis. The OF-spec-F/R primers used in the PCR did not react with the DNA of M. bilis or other metacercariae of trematodes parasitizing in the ide. The PCR assay with DNA of the red fox and different cestodes common for carnivores also proved to be negative. The primers are highly specific for the detection of O. felineus DNA while examining the sensitivity of OF-spec-F/R primers, the DNA of O. felineus could still be detected till 2 ng.
The specificity and sensitivity of the MB-spec-F/R primers for the detection of M. bilis DNA was tested in the same way as described for the OF-spec-F/R primers. In all cases, there was a fragment of approximately 167 bp. When testing the MB-spec-F/R primers with DNA from the trematode O. felineus, different metacercariae found in L. idus, and different cestodes found in the red fox, the results also proved to be negative. Nonspecific reaction was detected only when DNA of M. xanthosomus metacercariae was used that might be explained by the fact that M. bilis and M. xanthosomus belong to the same genus. The sensitivity of MB-spec-F/R primers with M. bilis DNA showed that it could still be detected till 1 ng.
CONCLUSIONS
Multiplex PCR technique is a useful tool to solve the epidemiological tasks using different sources of clinical probes including human, mammals or such intermediate hosts as fish. In particular, this is important for specifying the pathogenic species of opisthorchiidae trematodes throughout their overlapping ranges. Likewise, our PCR assay demonstrated no cross-reaction with the DNAs of two opisthorchiidae trematodes or with other helminthes. So, it can be also applied for opisthorchiidae-like eggs detection in the feces and bile of humans and wild/domestic mammals.
ACKNOWLEDGEMENT
The authors are grateful to Aibek Zhumalin (S. Seifullin Kazakh Agrotechnical University, Nur-Sultan, Kazakhstan) who helped to order specific primers and conducted sequencing of PCR-amplified DNA fragments and Lyudmila Lider (S. Seifullin Kazakh Agrotechnical University, Nur-Sultan, Kazakhstan) for their technical support in fish and animals testing for the presence of opisthorchiid trematodes.
FUNDING
All authors agree to the conditions outlined in the copyright assignment form included. This work was supported by the Ministry of Education and Science of the Republic of Kazakhstan, grant numbers AP05131132, 2018–2020.
Authors Contribution
VS and AK conceived the project, researched data, analyzed data and drafted the manuscript. AB, AS, and SB researched the data and reviewed and edited the manuscript. All authors read and approved the final draft of the manuscript.
Conflict of interest
The authors have declared no conflict of interest.
REFERENCES