Advances in Animal and Veterinary Sciences
Research Article
Validation of One-step Multiplex RT-PCR for Diagnosis of Respiratory Viruses Coinfections in Chickens
Marwa Hamed I. Salem1*, Naglaa M. Hagag2, Ahmed A.H. Ali3, Mohammed Saed M. El-Shahidy4
1Department of Virology, Zagazig Branch, Animal Health Research Institute, Dokki, Giza, Egypt. 2RLQP, Animal Health Research Institute, Dokki, 12618, Giza, P.O. Box 246, Egypt. 3Department of Virology, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt. 4Department of Virology, Faculty of Veterinary Medicine, Suez Canal University, Ismailia, Egypt.
Abstract | Avian influenza (AI), Infectious Bronchitis (IB), and Newcastle disease (ND) are frequently occurring respiratory infections of the chicken. These are highly fatal diseases caused by three devastating viruses in chicken farms in Egypt. Clinical diagnosis, serological tests, and virus isolation are the classical methods currently used for the characterization of these viruses. However, these methods have several concerns related to sensitivity and specificity that can best be managed with rapid diagnostic molecular techniques. Originating from five governorates (Ismailia, Sharkia, Gharbia, Dakahleia and Matrooh), a total of 138 nasal swabs and 144 tissue pools were collected from ten broiler chicken flocks with a history of severe respiratory diseases and tested by quantitative reverse transcriptase-polymerase chain reaction (RT-qPCR). The RT-qPCR positive samples were subjected to multiplex RT-PCR for the diagnosis of three major avian infections (AI, IB, and ND). The results showed that multiplex RT-PCR is sensitive for both uniplex and mRT-PCR as results showed that The lower limit of detection (LOD) for NDV was found to be 10-3, 10-4 for IBV, and 10-5 for AIV. This LOD was found to be corresponding to 15, 14, and 19 picograms for the RNA extracted from the reference strains. Also the results showed that multiplex RT-PCR is very suitable as a routine laboratory test for rapid and specific detection of co-infections in field samples, as it showed that single virus infection is prominent than double and triple virus infections with a percentage of 75%, while double virus co-infection is more frequent than triple virus co-infection with a percentage of 15% and 10 % respectively.
Keywords | Co-infection, Avian influenza virus, Infectious bronchitis virus, Newcastle disease virus, RT-qPCR, Multiplex RT-PCR
Received | June 10, 2020; Accepted | June 25, 2020; Published | July 10, 2020
*Correspondence | Marwa Hamed I. Salem, Department of Virology, Animal Health Research Institute, Zagazig, 44516, Egypt; Email: dr_marwa.vet2011@yahoo.com
Citation | Salem MHI, Hagag NM, Ali AAH, El-Shahidy MSM (2020). Validation of one-step multiplex RT-PCR for diagnosis of respiratory viruses coinfections in chickens. Adv. Anim. Vet. Sci. 8(s1): 62-67.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.s1.62.67
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 Salem et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Caused by viruses, respiratory diseases are the most devastating in the poultry industry due to their major economic losses. In most cases, there is more than one virus involved simultaneously in respiratory viral diseases (Malik et al., 2004). Among many avian respiratory viruses, AIV, IBV, and NDV are the most important co-infected viruses of poultry in Egypt (Ali and Reynolds, 2000). AI, caused by influenza A viruses of the Orthomyxoviridae family, infects a wide range of birds and cause respiratory or systemic diseases that vary in severity and cause heavy losses to the poultry industry worldwide (Aly et al., 2008) including Egyptian provinces (Dabbour, 2015). AIV is subtyped according to their surface glycoproteins named hemagglutinin (H) and neuraminidase (N). Currently, there are 18 H glycoproteins (H1 to H18) and 11 N glycoproteins (N1 to N11) (CDC, 2018). The IB, caused by IB virus (IBV), results in high mortality, affects weight gain and feed efficiency, and increased condemnations (Cavanagh and Gelb, 2008). The virus (IBV) undergoes mutations in chickens resulting in the emergence of new variant serotypes and genotypes that can escape vaccine immunization (Dolz et al., 2008). With the emergence of novel strains of IBV, besides an understanding of disease epidemiology and virus evolution, it is essential to rapidly detect and implement control measures (De-Wit, 2000). Similarly, caused by avian Paramyxovirus 1, ND is an important contagious disease worldwide (Cao et al., 2013), including Egypt. It is classified as a List A disease by OIE (OIE, 2012).
Replaced by the classical methods, molecular techniques are recently well-established in the virology laboratories for the diagnosis of viral infections. Widespread usage of polymerase chain reaction (PCR) as a rapid diagnostic technique has enhanced the capability of a laboratory for the diagnosis of any clinical disease. As a modification to the conventional PCR test, the use of multiplex primers has further enhanced the viral detection sensitivity (Alexander, 2000). Multiplex polymerase chain reaction (M-PCR) is the simultaneous detection of more than one virus in a tube reaction. The first report of multiplex PCR included the diagnosis of inherited genetic diseases (Dieffenbach et al., 1993). Further, it was proved to be a significant method for pathogen identification (Kalvatchev et al., 2004), where, along with a reduction in effort and time, it was found to be more sensitive and specific (Edwards and Gibbs, 1994).
A simultaneous coinfection of chickens with more than one virus has been evidenced in many chicken farms in Egypt. Therefore, the current study aimed to develop and validate multiplex reverse transcriptase-PCR (mRT-PCR) for detection and differential diagnosis of AIV, IBV and NDV.
Materials and Methods
Local virus strains
Local reference viruses (AIV with accession no. MH893738, KP209303 and MH762070 and NDV with accession no. KU365663 and IBV with accession no. KT832809) that were used as a positive control in the standardization and validation of multiplex RT- PCR are listed in Table 1. These viruses were isolated from Egyptian poultry farms and were characterized by RLQP (Reference Laboratory for Veterinary Quality Control on Poultry Production) -animal health research institute, Dokki, Egypt. Virus strains were propagated in 11-d-old SPF embryonated chicken eggs (ECEs) up to three passages for IBV and tested according to the method described previously (Beard, 1980; Swayne et al., 1998).
Clinical samples
A total of 138 nasal swabs and 144 tissue samples (brain, trachea, lung, liver, proventriculus, spleen, kidney, intestine, and cecal tonsils) were collected from broiler farms, originating from five Egyptian governorates (1- Ismailia, 2- Sharkia, 3- Gharbia, 4-Dakahleia, and 5- Matrooh (Figure 1)). Individual tissue samples for each bird were pooled in a tube and processed accordingly. All sampled chickens were suffering from severe respiratory illness, with mortalities ranging from 5 to 20%. Nasal swabs and tissue pools were prepared and processed according to the methods described previously (OIE, 2014).
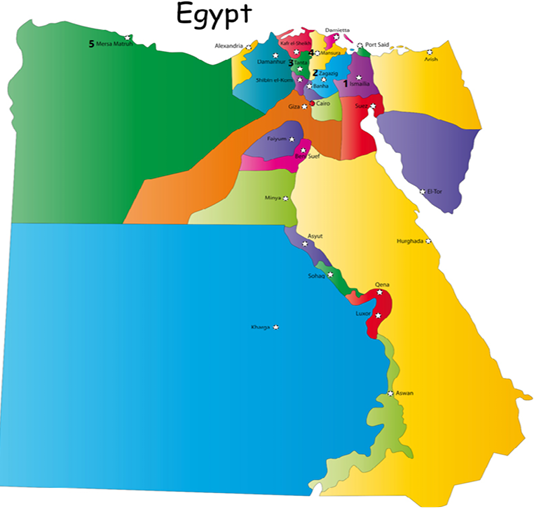
Figure 1: A map of Egypt showing sampling locations as 1- Ismailia, 2- Sharkia (Zagazig), 3- Gharbia (Tanta), 4-Dakahleia (Mansura), and 5- Matrooh.
RNA extraction
Total RNA was extracted from swabs, tissue pools, and reference virus isolates using Dynabeads® SILANE viral NA Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Primers and probes
Three sets of primers and probes were used for the detection of AIV, NDV, and IBV in the qRT-PCR and the mRT-PCR. Primers and Probes were specifically amplified targeting M, NP, F genes of AIV, IBV, and NDV respectively in qRT-PCR where primers amplified M, S1, F genes of AIV, IBV and NDV respectively in mRT-PCR. The sequence and length of amplified fragments of the primers and probes are listed in Tables 2 and 3. The sequences were obtained from published data in the literature and synthesized by Operon Biotechnologies (Huntsville, AL) (Naglaa et al., 2014; Gelb et al., 2005; Adzhar et al., 1997, Friedrich-Loeffler-Institute).
Table 1: Local reference virus strains and their accession number.
| Accession No | Reference strain | Virus |
| MH893738 | Influenza A virus (A/duck/Egypt/SS19/2017) | AI/H5N8 |
| KP209303 | (A/goose/Egypt/1439FAOS/2014(H5N1) | AI/H5N1 |
| MH762070 | Influenza A virus (A/chicken/Egypt/18558V/2018) | AI/H9N2 |
| KU365663 | NDV-31-EGYPT-NLQP-2014 | NDV |
| KT832809 | IBV-NLQP-15919F | IBV |
Table 2: Primer and Probe sequences used for qRT-PCR.
| IBV-F | 5-GCTTTTGAGCCTAGCGTT-3 |
| IBV-R | 5-GCCATGTTGTCACTGTCTATTG-3 |
| IBV-PROBE | 5-FAM-CACCACCAGAACCTGTCACCTC-BHQ1-3 |
| NDV-F | 5-TCCGGAGGATACAAGGGTCT-3 |
| NDV-R | 5-AGCTGTTGCAACCCCAAG-3 |
| NDV-PROBE (VFP-1) | 5-FAM-AGCGTTTCTGTCTCCTTCCTCCA-TAMRA-3 |
| sepro-01 | 5-AGA TGA GTC TTC TAA CCG AGG TCG-3 |
| Sepro-02 | 5-TGC AAA AACATC TTC AAG TCT CTG-3 |
| Sepro-probe | 5-FAM-TCAGGCCCCCTCAAAGCCGA-BHQ1-3 |
Table 3: Oligonucleotide primers used in RT-PCR and length of the amplified products.
| Virus | Primer name | Primer Sequence | Product size | Reference |
| NDV |
F330 R700 |
5-AGG AAG GAG ACA AAA ACG TTT TAT AGG-3 5-TCA GCT GAG TTA ATG CAG GGG AGG-3 |
400pb | |
| IBV |
HVR1-2-F HVR1-2-R |
5-GTKTACTACTACCARAGTGC -3 5- GAAGTGRAAACRAGATCACCATTTA -3 |
700pb | |
| AIV |
M-7FV2 M124R |
5-AAAGCAGGAAGATGTTGAAAGA-3 5-TGCAAAAACATCTTCAAGTCTCTG-3 |
124pb | (Friedrich-Loeffler-Institute) |
Codes for mixed bases positions: R, A/G; K, G/T.
Quantitative RT-PCR (qRT-PCR)
A one-step qRT-PCR was employed using INVITROGEN kits (Invitrogen, Carlsbad, CA, USA). following the protocol of the manufacturers. The cycling conditions were once each of 30 min at 48oC, and 10 min at 95oC once, followed by 40 cycles of each of 95oC for 15seconds, 60oC for 1 minute, where the fluorescence was recorded at 60oC.
Multiplex RT-PCR
Amplification of three RNA viruses (AIV, IBV, and NDV) were simultaneously performed to amplify M, S1, F genes of AIV, IBV, and NDV, respectively using SuperScriptTM III One-Step RT-PCR kit with Platinum®Tag (INVITROGEN). Multiplex RT-PCR was optimized and validated using local reference strains and the clinical samples. Several chemical and thermal conditions were evaluated, and the assay was optimized by adjusting primers concentrations, the thermal cycling temperatures, and the duration according to the method described previously (Markoulatos et al., 1999). The RT reaction was done at 48ºC for 30 minutes, along with an initial denaturation at 95 ºC for 3 minutes. This was followed by 35 cycles of each of 94 ºC for 20 seconds, 50ºC for 30 seconds, and 72 ºC for 20 seconds. The final extension was performed at 72 ºC for 5 minutes.
Results
qRT-PCR
Positive control viruses were used to determine the Ct value at which amplification of M, NP, and F gene begins, for AIV (A/chicken/Egypt/ 18558V /2018), for IBV (IBV-NLQP-15919F) and NDV (NDV-31Egypt-NLQP-2014). The amplification of the NP gene of reference IBV begins at 16.4 Ct value, while for the F gene of reference NDV and M gene of reference AIV, it begins at Ct 12 and 24, respectively. Nasal swabs and tissue pools of chickens infected with AIV, IBV, and NDV were examined by qRT-PCR using specific primers and probes for AIV, IBV, and NDV. All primer-probe pairs reacted only with their corresponding targets with mean Ct values ranging from 24.5-32.1 for AIV, 23.8-27 for NDV, and 26.3-31.2 for IBV.
Validation of monoplex (uniplex) and mRT-PCR
Optimization of RT-mix, Primer annealing temperature, and template concentrations:
RT-mix were optimized to 2.5µl, 5µl, 10µl, and 15µl in a total volume of 20µl. A maximum yield of specific PCR bands for the tested viruses was observed at an annealing temperature of 50oC and 56°C (Figure 2A). The optimum concentrations of the target RNA for Avian Influenza (15 μg/µl), Newcastle disease virus (20μg/μl), and Infectious Bronchitis virus (15 μg/μl) were determined.
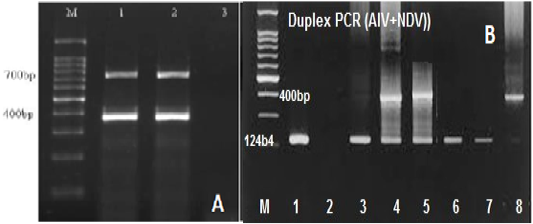
Figure 2: (A) Gel electrophoretic analysis of PCR product of duplex RT-PCR of reference IBV and NDV at Ta of 50oc; Lane M: DNA ladder; lanes 1and2: duplex RT-PCR products of IBV (700 bp), NDV (400bp); Lane 3: NTC. (B) Optimization of duplex RT-PCR in nasal swabs and tissue pools of chickens infected with AIV and NDV at 124 and 400bp respectively, lane 1 +ve AIV, lane 2 –ve, lane 3 +ve AIV, lane 4and5 +ve AIV+NDV, lane 6 +ve AIV, Lane 7 – ve, lane 8 Positive NDV.
The specificity of uniplex and duplex RT-PCR
The developed uniplex and mRT-PCR for reference strains of AIV, IBV, and NDV were affirmed to be particular by amplifying RNA fragments of three reference viruses. The assay produced amplicons tha were expected for each of the three viruses as 124bp for AIV, 700bp for IBV, and 400bp for NDV. No amplicon was detected by negative control or no template control (NTC).
The sensitivity of uniplex and mRT-PCR
The sensitivity of both uniplex and mRT-PCR was assessed using a 10-fold serial dilution of the cDNA of reference positive control extract. RNA of positive control for each virus was diluted (10-1 to 10-8), and each of the dilutions processed for uniplex and multiplex PCR. The lower limit of detection (LOD) was found to be 10-3 for NDV, 10-4 for IBV, and 10-5 for AIV. This LOD was found to be corresponding to 15, 14, and 19 picograms for the RNA extracted from the reference strains.
Optimization of multiplex RT-PCR in a clinical sample
An ability of multiplex RT-PCR to detect more than one virus template in a reaction was assessed in clinical samples (Figure 3). Tissue pools and swab samples proved to be positive for at least one of NDV or AIV or IBV by qRT- PCR. Results of the occurrence of co-infections in broiler farms among the five Egyptian provinces are presented in Table 4, showed that, single virus infection is prominent than double and triple virus infection as 60 samples out of 80 positives suffered of single virus infection with a total 75%, while the occurrence of the dual co-infection was more frequent than the triple one, where 15% of the sample had a dual occurrence of co-infection where, AIV with IBV mixed infection represents the most prominent than infection with IB and NDV with a total percentage of 7.5% and 5% respectively. The lowest double infection percentage observed when AIV mixed with NDV with a total percentage of 2.5% , while 10 % of the samples had a simultaneous occurrence of the three viruses in this study. (Figure 2B).
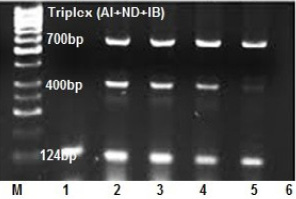
Figure 3: Optimization of triplex RT-PCR in nasal swabs and tissue pools of chickens infected with AIV, NDV and IBV at 124, 400 and 700 bp respectively. lane 1: AIV monoplex, lane 2and3and4: triplex (AI+IB+ND), lane 5: duplex (AIV+IBV), lane 6: NTC.
Table 4: Single, double and triple viral infections in 5 Egyptian province broiler farms.
| Govern. / total +ve | Single infection | Double infection | Triple infection | Total | |||
| No. | % | No. | % | No. | % | ||
| Sharkia-Swabs (11) | 6 | 54.54 | 3 | 27.27 | 2 | 18.18 | 11 |
| Tissue pools(11) | 8 | 72.72 | 2 | 18.18 | 1 | 0.1 | 11 |
| Dakahlia- swabs (8) | 7 | 87.5 | 1 | 12.5 | - | 00.00 | 8 |
| Tissue pools (8) | 7 | 87.5 | 1 | 12.5 | - | 00.00 | 8 |
| Gharbia- Swab (8) | 6 | 75 | 1 | 12.5 | 1 | 12.5 | 8 |
| Tissue pool (8) | 5 | 62.5 | 2 | 25 | 1 | 12.5 | 8 |
| Matrouh-Swab (4) | 4 | 100 | - | 00.00 | - | 00.00 | 4 |
| Tissue pool (4) | 4 | 100 | - | 00.00 | - | 00.00 | 4 |
| Ismailia- Swab (9) | 6 | 66.66 | 1 | 11.11 | 2 | 22.22 | 9 |
| Tissue pool (9) | 7 | 77.77 | 1 | 11.11 | 1 | 11.11 | 9 |
| Total (80) | 60 | 75 | 12 | 15 | 8 | 10 | 80 |
Discussion
Respiratory infections are of great concern around the world, which are frequently related to the simultaneous occurrence of infections (Yashpal et al., 2004). Coinfections of poultry complicate and hinder the identification and diagnosis (Costa-Hurtado et al., 2014). A co-infection in poultry with other respiratory viruses has a significant impact on disease pathogenicity, e.g., an occurrence of AIV along with NDV and IBV has a significant effect on the disease pathogenicity (Pan et al., 2012; Pu et al., 2012). Using molecular methods, for instance, conventional PCR, an individual detection of viruses from a co-infection is laborious, intensive, and expensive. This disadvantage is overcome by developing an mRT-PCR that uses more than one primer set for amplification of many genes in a single reaction (Elnifro et al., 2000). Indeed, mRT-PCR method amplifies many genes of several viruses simultaneously and, therefore, this method reduces the need for reagents and the personnel time (Renshaw et al., 2001).
For RT-PCR amplification, the target sequences selected were Matrix (M) gene for AI virus, Fusion Protein (F) gene for ND virus and Surface protein 1 (S1) for IB virus. First, the amplification is conducted as monoplex RT-PCR in individual reaction for different reaction and different amplification condition, then to do more effective, efficient and easier rapid diagnostic techniques, this study has been developed a modification for the simultaneous detection of AI, ND and IB viruses in a single step multiplex RT-PCR reaction. In either monoplex or multiplex RT-PCR for amplification of viral genes, each primer set produced virus specific products of expected size of 124 bp for M gene of AI virus, 400 bp for F gene of ND virus and 700 bp for S1 gene of IB virus respectively.
The results of multiplex PCR for these three respiratory viruses suggest that multiplex PCR is able to detect and differentiate the presence of these respiratory viruses in the clinical samples (Table 4). A multiplex PCR that simultaneously detects and differentiates the three major respiratory viruses of chicken will be highly advantageous to the poultry industry, particularly in Egypt. Rapid but specific detection without the need for subculture in host systems would greatly aid the diagnosis and control of outbreaks. A multiplex PCR system will be more economical and will require less time than a single PCR for each of these three avian respiratory pathogens.
Conclusions
A multiplex RT-PCR assay can be used for the simultaneous detection of more than one virus in one sample in a short time. The assay has found to be specific, sensitive and requires fewer reagents and time than conventional PCR. Also, the assay can be used to diagnose difficult-to-culture viruses from clinical samples such as IBV. Further studies are necessary to ascertain study outcomes and its potential application on other poultry viruses of immense importance.
Acknowledgements
This work was supported by the Faculty of Veterinary Medicine, Zagazig University, Egypt.
Authors Contribution
All authors contributed equally to this work.
Conflicts of Interest
The authors have declared no conflict of interest.
References





