Advances in Animal and Veterinary Sciences
Research Article
Antimicrobial Resistance Genes Associated with Enterococci from Poultry in Egypt, First Reporting of mecA in Enterococcus from Poultry Source
Wedad A. Ahmed1,2*, Helmut Hotzel1, Heinrich Neubauer1, Ashraf A. Abd El-tawab2, Herbert Tomaso1, Mona M. Sobhy3, Fatma I. El Hofy2
1Friedrich-Loeffler-Institute, Institute of Bacterial Infections and Zoonoses, Naumburger Str. 96a, 07743 Jena, Germany; 2Department of Microbiology, Faculty of Veterinary Medicine, Benha University, PO Box 13736, Moshtohor Toukh, Egypt; 3Department of Reproductive Diseases, ARRI, ARC, 12556 Giza, Egypt.
Abstract | Little information available for the Egyptian setting. 117 cloacal swabs were collected from diseased poultry located in 6 districts in Egypt. Isolates were identified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). The antibiotic susceptibility testing of Enterococcus faecium and Enterococcus gallinarum isolates against 22 antibiotics was performed with the MICRONAUT system for Gram-positive bacteria. The presence of 12 resistance-associated genes (tetK, tetL, tetM, tetS, tetO, erm(B), msrC, aac6-aph2, vanA, vanB, vanC1 and mecA) was investigated by PCR. Thirty one Enterococcus isolates have been detected, Enterococcus faecium (n=22) and Enterococcus gallinarum (n=9) strains were isolated. All Enterococcus faecium and Enterococcus gallinarum isolates were multidrug-resistant. More than 15% of Enterococcus faecium isolates were resistant to vancomycin. Resistance rates to other antibiotics ranged between 31.8% for teicoplanin to 86.4% for penicillin G, respectively. Enterococcus gallinarum isolates showed a resistance rate of 100% to cefoxitin, ceftaroline, clindamycin, daptomycin, erythromycin, gentamicin and oxacillin. The resistance genes most often found were msrC, erm(B), aac6-aph2 and mecA. The vanB gene was identified in two Enterococcus faecium isolates from which one was resistant to vancomycin whereas vanC1 was detected in 6 Enterococcus faecium and Enterococcus gallinarum isolates. Tetracycline resistance-associated genes (tetK, tetL, tetM, tetO and tetS) were found in 25 isolates whereas mecA was detected in nine isolates. The study is the first report for mecA gene in Enterococcus. The development of resistance to antibiotics in poultry production needs special attention and investigation. The usage of antimicrobials in Egypt should follow WHO and OIE recommendations.
Keywords | Enterococcus, VRE, Antibiotic resistance, mecA, Egypt
Received | December 22, 2019; Accepted | April 8, 2020; Published | May 12, 2020
*Correspondence | Wedad Ahmed. Friedrich-Loeffler-Institute, Institute of Bacterial Infections and Zoonoses, Naumburger Str. 96a, 07743 Jena, Germany; Email: drwedad_2009@yahoo.com
Citation | Ahmed WA, Hotzel H, Neubauer H, El-tawab AAA, Tomaso H, Sobhy MM, El-Hofy FI (2020). Antimicrobial resistance genes associated with enterococci from poultry in Egypt, first reporting of mecA in Enterococcus from poultry source. Adv. Anim. Vet. Sci. 8(6): 570-581.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.6.570.581
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 Ahmed et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Poultry has been reported to carry increasingly antibiotic-resistant bacteria in its digestive tract. In the past, enterococci of the gut of humans and animals were considered to be common and harmless with little or no clinical relevance (Fisher and Phillips, 2009; Marshall and Levy, 2011). Meanwhile, they are the second and third most important etiologic agents of urinary tract infections and nosocomial bacteremia in humans, respectively. In intensive care units enterococci may be found in 14.3% of patients (Papadimitriou-Olivgeris et al., 2014). The most common species isolated from clinical samples of humans are Enterococcus (E.) faecalis and E. faecium (Panesso et al., 2008).
One of the main reasons that enterococci can survive in the hospital environment is their intrinsic resistance to some antibiotics and their ability to acquire resistance. Previous studies have shown that the prevalence of antimicrobial resistance and virulence factors of human and animal Enterococcus isolates may vary according to geographical origin and antimicrobial regime (Kwon et al., 2012; Liu et al., 2013).
Vancomycin-resistant E. faecium is the second leading pathogen of the priority list of antimicrobial resistance (priority pathogens) published recently by WHO that are a major threat to public health (WHO, 2017).
In poultry, enterococci are frequently isolated in cases of endocarditis, arthritis, amyloidosis and various disorders (Kense and Landman, 2011). Enterococci can also cause food intoxication through production of biogenic amines (Asadian et al., 2016).
Consumption of antimicrobials is an important risk factor for colonization with multidrug-resistant enterococci because of the suppression of the competitive indigenous microbiota of the gastrointestinal tract. The increased number of gut enterococci frequently results in bloodstream infections (Ubeda et al., 2010). Enterococci are known to easily acquire and transfer virulence and antimicrobial resistance determinants via uptake of mobile genetic elements of commensals and pathogens alike (Oliveira et al., 2016).
Several studies have been conducted to determine the prevalence, antimicrobial and virulence mechanisms of enterococci isolated from broilers in different countries (Ngbede et al., 2017; Nowakiewicz et al., 2017). However, little information is available for the Egyptian setting. Therefore, this study investigates presence and antimicrobial resistance of Enterococcus species in poultry flocks.
Materials and Methods
Sample collection and cultivation of enterococci
Between January 2017 and November 2017, a total of 117 cloacal swabs were collected aseptically from diseased poultry with gastrointestinal disorders of 38 poultry farms (layers chicken, broilers, turkeys and ducks) located in 6 districts of Egypt (Qulyobia, Sharkia, Dakahlia, Damietta, Monofia and Gharbia governorates). The swabs were placed into micro-tubes containing sterilized phosphate-buffered saline. They were transported to the laboratory and immediately streaked out onto blood agar containing 5% sheep blood and incubated at 37.7°C for 48 h (Ulger et al., 2009).
Identification by MALDI-TOF MS
Isolates were identified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Bizzini et al., 2010). Briefly, bacteria from overnight cultures were suspended in 300 µl of bi-distilled water and mixed with 900 µl of ethanol (96% vol/vol; Carl Roth GmbH, Karlsruhe, Germany) for protein precipitation. After centrifugation for 5 min at 10,000 x g, the supernatant was removed and the pellet was re-suspended in 50 µl of 70% (vol/vol) formic acid (Sigma-Aldrich Chemie GmbH, Steinheim, Germany). Fifty microliters of acetonitrile (Carl Roth GmbH) were added, mixed and centrifuged for 5 min at 10,000 x g. One and a half microliter of the supernatant was transferred onto a MTP 384 Target Plate Polished Steel TF (Bruker Daltonik GmbH, Bremen, Germany). After air-drying the material was overlaid with 2 µl of a saturated solution of α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich Chemie GmbH) in a mix of 50% acetonitrile and 2.5% trifluoroacetic acid (Sigma-Aldrich Chemie GmbH). After air-drying spectra were acquired with an Ultraflex instrument (Bruker Daltonik GmbH). The instrument was calibrated with the IVD Bacterial Test Standard (Bruker Daltonik GmbH). Analysis was carried out with the Biotyper 3.1 software (Bruker Daltonik GmbH). Interpretation of results was performed according to the manufacturer’s recommendation: score of ≥ 2.3 represented reliable species level identification; score 2.0–2.29, probable species level identification; score 1.7–1.9, probable genus level identification, and score ≤ 1.7 was considered an unreliable identification (Lüthje et al., 2017).
Antibiotic susceptibility testing
The antibiotic susceptibility testing of all isolates was performed with the MICRONAUT system for Gram-positive bacteria using commercial 96-well microtiter plates (Merlin, Bornheim, Germany) according to the manufacturer´s recommendations. This system allowed the determination of minimum inhibitory concentrations (MICs) of 22 antimicrobial agents including ampicillin (β-lactam), cefoxitin (β-lactam; cephamycin), ceftaroline (cephalosporin 5th generation), clindamycin (lincosamide), daptomycin (cyclic lipopeptide), erythromycin (macrolide), erythromycin/clindamycin, fosfomycin (epoxide antibiotic), fusidic acid (steroide antibiotic), gentamicin (aminoglycoside), linezolid (oxazolidinone), moxifloxacin (fluorochinolone 4th generation), mupirocin, oxacillin (β-lactam), penicillin G (β-lactam), rifampicin (ansamycine), synercid (streptogramine), teicoplanin (glycopeptide), tigecycline (glycylcycline), trimethoprim/sulphamethoxazole (trimethoxybenzyl pyrimidine/ sulfonamide) and vancomycin (glycopeptide) in serial dilutions of the antibiotics. Bacteria grown overnight and suspended in NaCl solution (0.9%) to obtain a turbidity corresponding to a McFarland standard of 0.5 (Dr. Lange, CADAS photometer 30, Berlin, Germany). Three hundred microliters of the suspension were added to 11 ml of Mueller–Hinton broth (Oxoid Deutschland GmbH, Wesel, Germany) resulting in a concentration of approximately 106–107 colony forming units (cfu)/ml. In total, 100 µl of the inoculum were put in each well. After sealing the plates, they were incubated for 18 h to 24 h at 37°C. Reading of plates was done optically. Interpretation was carried out as recommended by the Clinical and Laboratory Standards Institute (CLSI, 2017).
Detection of resistance-associated genes
Genomic DNA was extracted from bacterial cultures using High Pure PCR Template Purification Kit (Roche Diagnostics, Mannheim, Germany) according to the instructions of the manufacturer. PCR amplifications of tetracycline resistance genes (tetK, tetL, tetM, tetS and tetO), erythromycin resistance gene (erm(B)), macrolide resistance gene (msrC), aminoglycoside resistance genes (aac6-aph2), vancomycin resistance genes (vanA, vanB and vanC1) and mecA gene were carried out using primers given in Table 1. PCR products were analyzed by electrophoresis on 2% agarose gel following staining with ethidium bromide and visualizing under UV.
For confirmation of PCR results, PCR products for all genes used were sequenced. Briefly, the bands of amplicons were cut out after electrophoresis and purified using the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) according to the manufacturer´s instructions. Cycle sequencing was done using the Big Dye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Darmstadt, Germany) according to instructions of the manufacturer. Sequencing products were analyzed with a Genetic Analyzer ABI Prism 3130 (Applied Biosystems), identification of the sequences was carried out by BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Results
Isolation and identification of Enterococcus species
Thirty-one Enterococcus isolates were isolated (Table 2). Using MALDI-TOF MS two species were identified representing 28 E. faecium (52.8%) and 9 E. gallinarum (17.0%) strains.
Antimicrobial susceptibility profiles
Antimicrobial resistance profiles of 22 E. faecium and 9 E. gallinarum isolates could be determined (Table 3a) and (Table 3b). These 31 isolates were multidrug-resistant which means that they were resistant to at least three antimicrobial agents of different classes. All E. faecium isolates were resistant to daptomycin, clindamycin, erythromycin and erythromycin/clindamycin. Four E. faecium isolates were resistant to vancomycin. Resistance rates to other antibiotics ranged between 31.8% for teicoplanin and 86.4% for penicillin G and cefoxitin.
Table 1: Primer and their sequences used for the detection of antibiotic resistance-associated genes in Enterococcus species.
| Gene | Primer sequences (5´-3´) | Expected amplicon size (bp) | Reference |
|
vanA |
F: ATG AAT AGA ATA AAA GTT GCA ATA R: CCC CTT TAA CGC TAA TAC GAT CAA |
1030 |
Getachew et al., 2012 |
|
vanB |
F: AAG CTA TGC AAG AAG CCA TG R:CCG ACA AAA TCA TCC TC |
536 |
Getachew et al., 2012 |
|
vanC1 |
F:GGA ATC AAG GAA ACC TC R:CTT CCG CCA TCA TAG CT |
822 |
Ünal et al., 2017 |
|
erm(B) |
F:GAA AAG GTA CTC AAC CAA ATA R:AGT AAC GGT ACT TAA ATT GTT TAC |
639 |
Sutcliffe et al., 1996 |
|
msrC |
F: AAG GAA TCC TTC TCT CTC CG R: GTA AAC AAA ATC GTT CCC G |
342 |
Werner et al., 2001 |
|
tetK |
F: TCG ATA GGA ACA GCA GTA R: CAG CAG ATC CTA CTC CTT |
169 |
Ng et al., 2001 |
|
tetL |
F: TCG TTA GCG TGC TGT CAT R: GTA TCC CAC CAA TGT AGC CG |
267 |
Ng et al., 2001 |
|
tetM |
F: GTG GAC AAA GGT ACA ACG AG R: CGG TAA AGT TCG TCA CAC AC |
406 |
Ng et al., 2001 |
|
tetO |
F: AACTTA GGC ATT CTG GCT CAC R: TCC CAC TGT TCC ATA TCG TCA |
515 |
Ng et al., 2001 |
|
tetS |
F: TGG AAC GCC AGA GAG GTA TT R: ACA TAG ACA AGC CGT TGA CC |
660 |
Aarestrup et al., 2000 |
|
aac6-aph2 |
F: CCA AGA GCA ATA AGG GCA TA R: CAC TAT CAT AAC CAC TAC CG |
219 |
van Asselt et al., 1992 |
|
mecA |
F: TCC AGA TTA CAA CTT CAC CAG G R: CCA CTT CAT ATC TTG TAA CG |
161 |
Pedroso et al., 2018 |
Table 2: Enterococcus species isolated from cloacal swabs of different poultry species after cultivation and MALDI-TOF MS identification.
| Source | No. of samples | No. of Enterococcus positive samples (%) | No. of isolates of different Enterococcus species identified in Enterococcus positive samples (%) | |
|
E. faecium |
E. gallinarum | |||
| Layer chickens | 54 | 15 (27.7) | 11 (73.3) | 4 (26.6) |
| Broiler chickens | 31 | 8 (25.8) | 5 (52.5) | 3 (37.5) |
| Turkeys | 4 | 3 (75.0) | 3(100) | - |
| Ducks | 28 |
5(17.8) |
3(60.0) | 2 (40.0) |
| Total | 117 | 31 (26.4) | 22 (70.9) | 9 (29.0) |
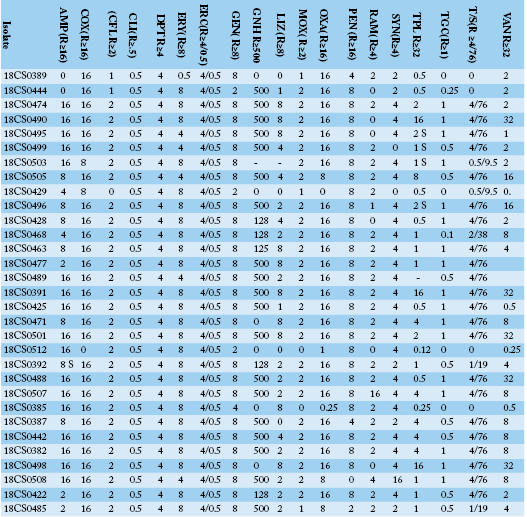
Table 3a: Antimicrobial resistance profiles, MIC values for each individual isolate of Enterococcus faecium and Enterococcus gallinarum isolates from Egyptian poultry.
AMP: ampicillin; COX: cefoxitin; CFL: ceftaroline; CLI: clindamycin; DPT: daptomycin; ERY: erythromycin; ERC: erythromycin/clindamycin; GEN: gentamicin; GNH: gentamicin high level; LIZ: linezolid; MOX: moxifloxacin; OXA: oxacillin; PEN: penicillin G; RAM: rifampicin; SYN: synercid, TPL: teicoplanin; TGC: tigecycline; T/S: trimethoprim/sulphamethoxazole; VAN: vancomycin.
Table 3b: Antibiotic resistance of Enterococcus faecium and Enterococcus gallinarum isolates from Egyptian poultry.
| Antibiotic | Class | Enterococcus faecium (n=22) | Enterococcus gallinarum (n=9) | ||||||
| S | I | R |
Resistance rate (%) |
S | I | R |
Resistance rate (%) |
||
| Ampicillin |
β-Lactam |
7 | 2 | 13 | 59.1 | 4 | 2 | 3 | 33.3 |
| Cefoxitin |
β-Lactam; cephamycin |
2 | 1 | 19 | 86.4 | 0 | 0 | 9 | 100 |
| Ceftaroline |
Cephalosporin 5th generation |
3 | 1 | 18 | 81.8 | 0 | 0 | 9 | 100 |
| Clindamycin | Lincosamide | 0 | 0 | 22 | 100 | 0 | 0 | 9 | 100 |
| Daptomycin | Cyclic lipopeptide | 0 | 0 | 22 | 100 | 0 | 0 | 9 | 100 |
| Erythromycin | Macrolide | 0 | 0 | 22 | 100 | 0 | 0 | 9 | 100 |
| Erythromycin/ Clindamycin | Macrolide/ Lincosamide | 0 | 0 | 22 | 100 | 0 | 0 | 9 | 100 |
| Gentamicin | Aminoglycosides | 6 | 0 | 16 | 72.7 | 0 | 0 | 9 | 100 |
| Gentamicin high level | Aminoglycosides | 10 | 0 | 12 | 54.5 | 5 | 0 | 4 | 44.4 |
| Linezolid | Oxazolidinone | 10 | 4 | 8 | 36.4 | 6 | 1 | 2 | 22.2 |
| Moxifloxacin |
Fluorochinolone 4th generation |
5 | 2 | 15 | 68.2 | 2 | 0 | 7 | 77.8 |
| Oxacillin |
β-Lactam |
4 | 1 | 17 | 77.3 | 0 | 0 | 9 | 100 |
| Penicillin G |
β-Lactam |
3 | 0 | 19 | 86.4 | 1 | 0 | 8 | 88.9 |
| Rifampicin | Ansamycine | 9 | 2 | 11 | 50.0 | 1 | 0 | 8 | 88.9 |
| Synercid | Streptogramine | 1 | 5 | 16 | 72.7 | 2 | 0 | 7 | 77.8 |
| Teicoplanin | Glycopeptide | 8 | 7 | 7 | 31.8 | 8 | 0 | 1 | 11.1 |
| Tigecycline | Glycylcycline | 6 | 5 | 11 | 50.0 | 2 | 4 | 3 | 33.3 |
| Trimethoprim/ Sulphamethoxazole | Dihdrofolatreductase/ Sulfonamide | 5 | 2 | 15 | 68.2 | 2 | 1 | 6 | 66.7 |
| Vancomycin | Glycopeptide | 14 | 4 | 4 | 18.2 | 8 | 0 | 1 | 11.1 |
All E. gallinarum isolates showed resistance to cefoxitin, ceftaroline, clindamycin, daptomycin, erythromycin, erythromycin/clindamycin, gentamicin and oxacillin. One was resistant to vancomycin.
Detection of antibiotic resistance determinants in enterococci
Detection of resistance-associated genes showed the presence of several of these genes in the 31 phenotypically resistant enterococci isolates (Table 4). The most often identified resistance genes were msrC responsible for macrolide resistance (n=20) and erm (B) associated with erythromycin resistance (n=20). The aac6-aph2 genes connected with aminoglycoside resistance and tetL associated with tetracycline resistance were also detected (n=17 each).
Vancomycin resistance genes were found in 14 isolates. The vanB gene was identified in two E. faecium isolates from which one was resistant to vancomycin. The vanC1 gene was detected in four E. faecium isolates. None of these isolates was phenotypically resistant to vancomycin. Six E. gallinarum isolates harbored the vanC1 gene but only one of them was phenotypically resistant.
Genes corresponding to tetracycline resistance (tetK, tetL, tetM, tetS and tetO) were found in 25 isolates. The tetL gene was detected in 18 isolates (13 E. faecium and 5 E. gallinarum) followed by 16 tetM-carrying isolates (12 E. faecium and 4 E. gallinarum). Phenotypic resistance to tetracycline was not found even in isolates which harboured three of the tet genes (tetL, tetM, tetS). tetK was detected in five isolates. One of them was identified in an E. gallinarum isolate. One E. faecium isolate harboured the tetS gene.
The mecA gene corresponding to β-lactam resistance was detected in seven E. faecium and two E. gallinarum by PCR. Five out of 28 E. faecium isolates were phenotypically resistant to β-lactam antibiotics like ampicillin, oxacillin and penicillin G. E. gallinarum isolates showed also phenotypically resistance to β-lactam antibiotics.
To confirm the PCR results concerning mecA gene amplicons were sequenced. The sequence of PCR products was identical with GenBank entry (Acc. No. MK991791).
Discussion
Antimicrobial resistance in enterococci is not only of major concern in the clinical setting of hospitals. Bacteria may also affect animal health or may contaminate food of animal origin (Silva et al., 2012). The rising number of infections in humans caused by resistant bacteria that
Table 4: Genotypic and phenotypic resistance profiles of Enterococcus faecium and Enterococcus gallinarum isolated from different poultry species in Egypt.
| Source | Isolate | Species | Resistance-associated gene | Phenotypic resistance |
| Layer chickens | 18CS0389 | E. faecium |
erm(B), tetL, mecA |
COX, CLI, DPT, RAM |
| 18CS0444 | E. faecium |
erm(B), msrC, aac6-aph2, tetS, tetL, tetM |
COX, CLI, DPT, ERY, ERC, GNH, PEN | |
| 18CS0474 | E. faecium |
erm(B) |
AMP, COX, CFL, CLI, DPT, ERY, ERC, GEN, GNH, LIZ, MOX, OXA, PEN, RAM, SYN, TGC, T/S | |
| 18CS0490 | E. faecium |
erm(B), msrC, aac6-aph2, tetL, tetM |
AMP, COX, CFL, CLI, DPT, ERY, ERC, GEN, GNH, LIZ, MOX, OXA, PEN, SYN, TPL, TGC, T/S, VAN | |
| 18CS0495 | E. faecium |
msrC, vanC1, tetK, tetL, tetM, mecA |
AMP, COX, CFL, CLI, DPT, ERY, ERC, GEN, GNH, LIZ, MOX, OXA, PEN, SYN, TGC, T/S | |
| 18CS0499 | E. faecium |
msrC, vanB, aac6-aph2, tetL, tetM, mecA |
AMP, COX, CFL, CLI, DPT, ERY, ERC, GEN, GNH, MOX, OXA, PEN, SYN, T/S | |
| 18CS0503 | E. faecium |
erm(B), msrC, vanC1, aac6-aph2, tetK, mecA |
AMP, CFL, CLI, DPT, ERY, ERC, OXA, PEN, RAM, | |
| 18CS0505 | E. faecium |
erm(B), msrC, tetK |
COX, CFL, CLI, DPT, ERY, ERC, GEN, GNH, MOX, OXA, PEN, SYN, TPL, T/S | |
| 18CS0429 | E. faecium |
erm(B), msrC, tetL |
CLI, DPT, ERY, ERC | |
| 18CS0496 | E. faecium |
erm(B), msrC, vanC1, aac6-aph2 |
COX, CFL, CLI, DPT, ERY, ERC, GEN, GNH, MOX, OXA, PEN, SYN, TGC, T/S | |
| 18CS0428 | E. faecium |
tetK, mecA |
COX, CLI, DPT, ERY, ERC, GEN, MOX, OXA, PEN, SYN, TGC, T/S | |
| 18CS0468 | E. gallinarum |
msrC, vanC1, aac6-aph2, tetM |
COX, CFL, CLI, DPT, ERY, ERC, GEN, MOX, OXA, PEN, RAM. SYN | |
| 18CS0463 | E. gallinarum |
vanC1, tetM |
COX, CFL, CLI, DPT, ERY, ERC, GEN, LIZ, MOX, OXA, PEN, RAM, SYN, T/S |
|
| 18CS0477 | E. gallinarum |
erm(B), aac6-aph2, tetL, tetM |
COX, CFL, CLI, DPT, ERY, ERC, GEN, LIZ, MOX, OXA, PEN, RAM, SYN, TGC, T/S | |
| 18CS0489 | E. gallinarum |
msrC, aac6-aph2, tetL |
AMP, COX, CFL, CLI, DPT, ERY, ERC, GEN, GNH, MOX, OXA, PEN, RAM, SYN, T/S | |
| Broiler chickens | 18CS0391 | E. faecium |
erm(B), msrC, tetL |
AMP, COX, CFL, CLI, DPT, ERY, ERC, FUS, GEN, GNH, MOX, OXA, PEN, RAM, SYN, TPL, TGC, T/S, VAN |
| 18CS0425 | E. faecium |
erm(B), msrC, tetL, tetM |
AMP, COX, CFL, CLI, DPT, ERY, ERC, FUS, GEN, GNH, MOX, OXA, PEN, RAM, SYN, TGC, T/S |
|
| 18CS0471 | E. faecium |
erm(B), msrC, aac6-aph2, tetL, tetM |
COX, CFL, CLI, DPT, ERY, ERC, GEN, LIZ, MOX, OXA, PEN, RAM, SYN, TPL, TGC, T/S | |
| 18CS0501 | E. faecium |
msrC, tetL, tetM |
COX, CFL, CLI, DPT, ERY, ERC, GEN, LIZ, MOX, OXA, PEN, RAM, SYN, TGC, T/S, VAN |
|
| 18CS0512 | E. faecium |
msrC |
AMP, CFL, CLI, DPT, ERY, ERC, PEN | |
| 18CS0392 | E. gallinarum |
erm(B) |
COX, CFL, CLI, DPT, ERY, ERC, GEN, MOX, OXA, PEN, RAM | |
| 18CS0488 | E. gallinarum |
vanC1, aac6-aph2, tetL, mecA |
AMP, COX, CFL, CLI, DPT, ERY, ERC, FUS, GEN, GNH, MOX, OXA, PEN, RAM, SYN, TPL, TGC, T/S, VAN |
|
| 18CS0507 | E. gallinarum |
erm(B), vanC1, aac6-aph2 |
AMP, COX, CFL, CLI, DPT, ERY, ERC, GEN, GNH, MOX, OXA, PEN, RAM, SYN, TGC, T/S | |
| Turkeys | 18CS0385 | E. faecium |
erm(B), msrC, vanC1, aac6-aph2, tetM, mecA |
AMP, COX, CFL, CLI, DPT, ERY, ERC, LIZ, PEN, RAM, SYN |
| 18CS0387 | E. faecium |
erm(B), msrC, aac6-aph2, tetM |
COX, CFL, CLI, DPT, ERY, ERC, GEN, GNH, MOX, OXA, RAM, T/S | |
| 18CS0442 | E. faecium |
erm(B), msrC, vanC1, tetL, tetM, mecA |
AMP, COX, CFL, CLI, DPT, ERY, ERC, GEN, GNH, MOX, OXA, PEN, RAM, SYN, T/S | |
| Ducks | 18CS0382 | E. faecium |
aac6-aph2 |
AMP, COX, CFL, CLI, DPT, ERY, ERC, GEN, GNH, OXA, PEN, RAM, SYN, TPL, T/S |
| 18CS0498 | E. faecium |
msrC, vanB, aac6-aph2, tetL, tetM, mecA |
AMP, COX, CFL, CLI, DPT, ERY, ERC, GEN, LIZ, MOX, OXA, PEN, SYN, TPL, TGC, T/S, VAN | |
| 18CS0508 | E. faecium |
erm(B), msrC, vanC1, aac6-aph2, tetK, tetL, tetM |
AMP, COX, CFL, CLI, DPT, ERY, ERC, GEN, LIZ, MOX, OXA, PEN, SYN, TPL, TGC, T/S | |
| 18CS0422 | E. gallinarum |
erm(B), vanC1, aac6-aph2, tetL |
COX, CFL, CLI, DPT, ERY, ERC, GEN, OXA, PEN, RAM, SYN, T/S | |
| 18CS0485 | E. gallinarum |
erm(B), msrC, vanC1, aac6-aph2, tetL, tetM, tetO |
COX, CFL, CLI, DPT, ERY, ERC, GEN, GNH, OXA |
AMP: ampicillin; COX: cefoxitin; CFL: ceftaroline; CLI: clindamycin; DPT: daptomycin; ERY: erythromycin; ERC: erythromycin/clindamycin; GEN: gentamicin; GNH: gentamicin high level; LIZ: linezolid; MOX: moxifloxacin; OXA: oxacillin; PEN: penicillin G; RAM: rifampicin; SYN: synercid, TPL: teicoplanin; TGC: tigecycline; T/S: trimethoprim/sulphamethoxazole; VAN: vancomycin.
originate from animal reservoirs is of great concern. In fact, results from previous studies showed that transfer of resistance genes from enterococci of animal origin to enterococci in human beings occurred through the food chain (Lester et al., 2006).
In this study, the dominant Enterococcus species was E. faecium (70.9%) followed by E. gallinarum (29.0%) which is a similar to the findings of (Ünal et al., 2017) who isolated E. faecium (60.4%) and E. gallinarum (2.6%) from broiler samples. No other Enterococcus species were detected in this study which could be attributed to different origin and feed contamination (Butaye et al., 1999) E. faecium was also the most commonly isolated Enterococcus species from poultry cloacal swabs in Turkey (Dilik et al., 2010) In contrast, E. avium and E. gallinarum were found to be the most predominant Enterococcus species in pigeon and duck faeces samples in Egypt (Osman et al., 2019). Pigeon and duck faeces were collected in Cairo city and poor neighborhoods while in the presented study samples were collected in farms of six governorates outside the Egyptian metropolis. Thus, differences concerning the origin of samples, housing, feeding, breeding but also host specificity may have influenced study outcome.
E. faecium and E. faecalis are the most predominant enterococci species causing human infections worldwide (Billington et al., 2014; Kajihara, 2015). They are also a main cause of healthcare-associated infections (Ben Sallem, 2016). These two species have developed resistance to a wide variety of clinically important antibiotics (Ngbede et al., 2017; Ünal et al., 2017; Bertelloni et al., 2015; Kim et al., 2019). In Egypt, another often ignored but critical circumstance is the uncontrolled discharge of large amounts of pharmaceutical waste containing active compounds from antibiotic manufacturing plants into rivers and the soil environment. This practice contributes to the emergence of antibiotic-resistant organisms resulting in considerable hazard to public health (Grenni et al., 2018). Thus, the high rates of antibiotic resistance found in this study may be caused by uncontrolled use of antibiotics for therapeutic or prophylactic purposes. It is noteworthy, that antibiotics are still used as growth promoters included in feed for poultry.
In the present study, four E. faecium and one E. gallinarum strains showed resistance to vancomycin which is in accordance with results obtained by (Ünal et al., 2017) for broiler cloacal samples in Turkey. Vancomycin resistance was also detected in 10/153 (6.5%) of Enterococcus isolates originated from food samples which were collected in different supermarkets and groceries in Egypt (Raafat et al., 2016). (Osman et al., 2019) found vanB and vanC genes in 25.5% and 33.0% in enterococci isolates from poultry in Egypt, respectively. Those findings are was comparable to results found in this study. A similar frequency of resistance (23.1%) was found in Egyptian E. faecium isolates from hospitals (Moemen et al., 2015).
The vanA gene could not be detected in any of the isolates as E. feacalis is the common carrier for this gene, while vanB and vanC1 genes were found in 14 out of 31 E. faecium and E. gallinarum isolates by PCR. Although vanC are intrinsic in E. gallinarum, unfortunately it was not detected in 3 of E. gallinarum isolates. Only two of them showed phenotypic resistance, so it is evident that there exists a discrepancy between phenotypic resistance and presence of resistance-associated genes.
Vancomycin resistance in our study reached an alarming rate as it is used for the treatment of enterococcal infections in humans in Egypt in contrast to the situation in the EU (Hao et al., 2016). In contrast to the EU, where the use of avoparcin which shows chemical similarity to vancomycin is forbidden in livestock, feeding avoparcin is widely used in Egypt as growth promoter and for prevention of necrotic enteritis in the poultry production, which may have led to an increased prevalence of vancomycin resistance in bacteria (Bager et al.,1997).
The resistance rates to antibiotics which are of high importance for treatment of humans e.g. daptomycin, linezolid and tigecycline were 100%, 36.4% and 50.0%, respectively. In contrast, (De Jong et al., 2018) detected absence or low levels of resistance to these antibiotics. These results are alarming, because multidrug-resistance in enterococci in poultry isolates is very likely to spill over to the human population via food or intensive contact in farming. Egyptian doctors admitted hospital hygiene failure and overuse of antibiotics as major causes for the high rate of antimicrobial resistance in Egypt (Saied, 2006).
The highest frequencies of resistance were found to clindamycin, daptomycin, erythromycin and erythromycin/clindamycin. A finding which is similar to that of (Osman et al., 2019) nearly all of their isolates were resistant to clindamycin and erythromycin. The described resistance rates were higher than those obtained by (Aslantas, 2019). In that study resistance to erythromycin was the most common (77.1%).
The msrC gene as well as the erm(B) gene both associated with macrolide/erythromycin resistance were found in 67.7% and 64.5% of isolates, respectively, rates that were lower than those reported by (Nowakiewicz et al., 2017; Kim et al., 2019) They identified the erm(B) gene in 96.0% and 98.4% of their isolates, respectively. These genes are located on mobile genetic elements like Tn917, Tn551 and Tn3871 and can be easily transferred horizontally (Shaw and Clewell, 1985; Jurado-Rabadan et al., 2014).
Resistance of enterococci to tetracycline is commonly mediated by tet genes (tetK, tetL, tetM, tetS and tetO). Some isolates carried one or more of these genes, however, phenotypic resistance to tetracycline could not be detected. In contrast to this study (Kim et al., 2019) identified tetL in 89.3% and tetM in 95.3% of isolates from chicken meat, respectively. The high resistance rates in Egypt can be explained by the frequent use of tetracycline in both, human and animal medicine for treatment of diseases but also for prophylaxis. Especially in animal and poultry production the easily affordable tetracyclines are used without microbiological identification and antibiogram of the causative pathogen i.e. cheap (Haggag et al., 2018).
Although tetracycline and erythromycin are not the drugs of choice for treatment of enterococcal infections, resistance to them is still of great clinical importance because they are effective for the treatment of other bacterial infections (Arias et al., 2010). Therefore, the ease with which enterococci can disseminate erythromycin and tetracycline resistance traits to other bacteria raises serious clinical and public health concern (Hammerum, 2012).
Resistance to aminoglycosides represented by gentamicin was detected phenotypically in 72.7% of E. faecium and all E. gallinarum isolates. This rate was higher than those reported by other researchers (Ngbede et al., 2017). High level gentamicin resistance (HLGR) is clinically important, because a combination of gentamicin with cell wall-active antibiotics (e.g. β-lactams, vancomycin) has been used for treatment of severe enterococcal infections (e.g. endocarditis). HLGR was detected in 54.5% of E. faecium and 44.4% of E. gallinarum isolates which is higher as the rate was reported by (Aslantas, 2019) with 14% HLGR isolates. Resistance to aminoglycosides is mediated by the aac6-aph2 genes and was detected in 54.8% of isolates in which all of them showed phenotypic aminoglycoside resistance. A lower rate of aac6-aph2 was reported by (Padmasini et al., 2014) who found 38.2% of clinical isolates harbouring these genes.
Resistance to β-lactam antibiotics (ampicillin, cefatoxine, oxacillin and penicillin G) for both, E. faecium and E. gallinarum, was high in general and excelled the rates given by a previous report from (Ngbede et al., 2017) who found 45.1% of resistant enterococci isolated from poultry and cattle farms in Nigeria. Resistance to β-lactam antibiotics is an important problem of therapeutic medications, especially when associated with high levels of resistance to ami-noglycosides and glycopeptides as found in our study. Multiple mutations in the active site of the pbp5 gene are common causes for high-level resistance instead of over-expression of PBP5 protein as shown by other authors (Hsieh et al., 2006; Jureen et al., 2004).
Penicillin-resistance detected by (Gousia et al., 2015) was lower than in this study, 41% vs. 29%. Although mecA is mainly reported in staphylococci (Hanssen et al., 2004; El-Kharroubi et al., 1991) has detected mecA homologue in Enterococcus hirae that led to the deduction that mecA also occurs in non-staphylococcal genera (Fitzgerald et al., 2003). The mecA gene associated with resistance to methicillin/oxacillin was detected in nearly 29% of isolates. All of them showed phenotypic resistance to one or more antibiotics of the β-lactam group. This is the first description of the mecA gene in Enterococcus species isolated from poultry and our results have been confirmed by sequencing PCR product for mecA gene. This gene was detected in enterococci isolated from soft cheese (Resende et al., 2018) and E. faecalis isolated from surface water (Kassem et al., 2008). Transfer of mecA gene between Enterococcus isolates is mainly due to acquiring of plasmid and or transposons (Tang et al., 2014).
It has to be noted that Enterococcus species can acquire and spread genes responsible for antimicrobial resistance at much higher rates than Gram-negative ones (Chajęcka-Wierzchowska et al., 2015).
The trimethoprim/ sulphamethoxazole resistance was in E. faecium and E. gallinarum almost similar to that reported by (Stepien-Pysniak et al., 2016). They reported a resistance rate of 88% of poultry isolates. This finding may be caused by the fact that trimethoprim/sulphonamides are used regularly for prophylaxis and treatment of poultry and animal coccidiosis.
In Egypt the high level of resistance to above mentioned antimicrobials in comparison to other countries is possibly the result of transmission of genes between isolates of animal and human origin. Enterococci and fecal-derived bacteria are often present in river water at the same time as water is contaminated with sewage from both, humans and animals (Romanis, 2013). It has been shown that river water is indeed a major source of multidrug-resistant bacteria due to pollution by waste products of agriculture, animal excreta and raw sewage (Lupo et al., 2012). Thus, spread of enterococci and other bacteria carrying antibiotic resistance genes may regularly occur via the water of the River Nile that is used for all purposes without sterilization/decontamination e.g. drinking water for humans and animals but also irrigation purposes.
Conclusion
Enterococci have become high risk zoonotic pathogens which can cause life threatening health problems. Enterococci isolated from poultry, especially E. faecium are known to cause nosocomial diseases in humans. The characterization of those isolates will help to decision makers to decide on counter measures along the value chain and in clinical settings at the hospital. All E. faecium and E. gallinarum isolates were multidrug-resistant and may cause serious health problems. Vancomycin-resistance was found in several isolates and underlines the importance of prudent use of antibiotics. This study also considered as first report for mecA gene in Enterococcus isolates from poultry, thus provide information that enterococci from poultry are possible vectors for transmission of critical and risky resistance-associated genes.
In general, the use of antimicrobials in poultry and food production should be controlled and prudent. Use of last line agents as vancomycin in animal production should be prevented worldwide and meticulous control of levy quantifies also should be implemented in developing countries.
Acknowledgements
We would like to thank the Egyptian Cultural and Educational Office of the Arab Republic of Egypt in Germany and the Egyptian Ministry of Higher Education for financial support. The authors thank Peggy Methner for her technical assistance.
Authors Contribution
WA, HH and HT participated in the conception and design of the study. WA, MMS performed farm and laboratory work. WA, FIE, HH analyzed the data and drafted the manuscript. HN, AAA, HT participated in manuscript revision. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Funding
Not applicable.
Conflict of interest
The author reports no conflicts of interest in this work.
References





