Advances in Animal and Veterinary Sciences
Research Article
Virulence Profile of Enteropathogenic Escherichia Coli (Epec) Isolated From the Cases of Neonatal Calf Diarrhea
H. I. Hosein*, R.A. Azzam, M. Abo-Elwafa, Ahmed M.S. Menshawy, Sherin Rouby
Department of Veterinary Medicine, Faculty of Veterinary Medicine, Beni-Suef University, 62511, Egypt.
Abstract | A total of 14 (30.43%) diarrheic calves of 1-4 weeks out of 46 calves kept under poor hygienic conditions in a dairy farm were suffering clinically from acute diarrhea. Diarrheic calves showed fever, diarrhea, dehydration, pneumonia recumbence and death of two calves. Thirteen (92.85 %) isolates of E. coli out of the 14 collected rectal swaps of diarrheic calves were identified on bacteriological and molecular basis. The results of the amplification of phoAgene using PCR revealed that all E. coli isolates showed positive result for the presence of phoAgene, thus confirming their identity as E. coli. All tested E. coli isolates were positive for intimin (eae) A, attaching and effacing gene (gene species specific) for E. coli (100%). No isolate had shiga toxin1 (stx1), shiga toxin 2 (stx2), hemolysin (hylA), and E. coli enterotoxin genes heat stable enterotoxin (st) and heat labile enterotoxin (lt). The obtained results indicate the possible participation of pathogenic E. coli in calf diarrhea. EPEC represented 100% of the tested E. coli strains obtained from diarrheic calves. The obtained results indicated that EPEC infection is a major health problem among calves and suggests the significance of poor hygiene measures in the investigated farm the possible participation of calves in the zoonotic transmission of pathogenic E. coli.
Keywords | EPEC, Multiplex PCR, Virulence genes
Received | February 18, 2019; Accepted | June 03, 2019; Published | August 24, 2019
*Correspondence | H.I. Hosein, Department of Veterinary Medicine, Faculty of Veterinary Medicine, Beni-Suef University, 62511, Egypt; Email: abdelaalhosein@yahoo.com
Citation | Hosein HI, Azzam RA, Abo-Elwafa M, Menshawy AMS, Rouby S (2019). Virulence profile of enteropathogenic escherichia coli (epec) isolated from the cases of neonatal calf diarrhea. Adv. Anim. Vet. Sci. 7(9): 756-761.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.9.756.761
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Hosein et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Health of the newborn calves is a matter of balance concerning the pathogens, immune system of the newborn animal, environment and management system. Infection in newborn calves is multifactorial in nature and it seems to be the result of interaction between different factors that may contribute to the buildup of infection including the immunological, nutritional, and environmental factors, (Hosein, 2018).
Neonatal calf diarrhea is one of the most serious health problems in the livestock industry and an important cause of economic losses due to high morbidity and mortality rates, high treatment costs and low growth rate, (Al-Alo et al., 2018; Anderson et al., 2003; Bazeley, 2003). Diarrhea is the most important cause of losses in calves up to 30 days of age and still problematic, likely because of the multi-factorial nature of the disease, Gomez and Weese (2017). Difference between health and disease among new born calves is very often just a slight tip of a delicate balance that weighs calf and environmental factors with the pathogens to which the calf will be exposed.
E. coli an important enteric pathogen of bovine neonates, is established in intestines shortly after the birth and remains throughout life. It has been incriminated as a major cause of diarrhea characterized by progressive dehydration and death that may occur depending on the age of the calf when scour started and the pathotypes of E. coli involved (Nguyen et al., 2011). Several E. coli serotypes, causing morbidity and mortality, have been isolated from calves suffering from diarrhea (Wani et al., 2003). E. coli pathotypes that are incriminated in neonatal calf diarrhea include Enterotoxigenic (ETEC), Enteropathogenic (EPEC), Shigatoxigenic (STEC) which include subgroup Enterohemorrhagic (EHEC), Enteroinvasive (EIEC), Enteroaggregative (EAEC) and Enteroadherent E. coli (EAdEC), (Nagy and Fekete, 2005). Each of these pathotypes has different virulence factors that enable them to cause diarrhea by different mechanisms which allowing them to colonize, invade and survive the host immune defense leading to certain clinical manifestations (Mokady et al., 2005).
More recently, Attaching and effacing E. coli (AEEC) strains (E. coli strains which cause A/E lesions or at least carry the genes for this trait) such as EHEC and EPEC pathotypes have been implicated in diarrhea and dysentery, mostly in two to eight weeks old calves (Moxley and Smith, 2010; Shahrani et al., 2014). The pathogenicity of EPEC pathotype is attributed to the production of the protein Intimin (eae) which has the ability to form attaching and effacing (A/E) lesions on intestinal cells (Oswald et al., 2000).
Thiry et al. (2017) detected serogroup O80 in 40% of 104 EPEC isolates from calves with diarrhea from 42 farms in Belgium. These isolates harbored the eae-ξ and fliCH2 genes, similar to the O80 attaching-effacing Shigatoxigenic E. coli isolates found in humans in France and that might be emerging.They concluded that Enteropathogenic and attaching-effacing Shigatoxigenic Escherichia coli cause bloody diarrhea in humans and young calves.
Polymerase chain reaction (PCR) can be used for the diagnosis of E. coli infection with high accuracy and is considered as an easy tool for amplifying genes of interest specifically present in a target pathotype or sero-group (Mullis, 1990; Begum et al., 1993).
The present study was designed for identifying and genotyping of the pathogenic E. coli associated with an outbreak of neonatal calf diarrhea through detection of genes encoding virulence factors using PCR and studying of some epidemiological aspects of E. coli infection associated with neonatal calf diarrhea.
Material and Methods
Area of Study
The present work was conducted on a dairy cattle private farm located in Beni-Suef Governorate, Egypt during December 2017- March 2018).
Animals
A total of 14 (30.43%) diarrheic calves of 1-4 weeks out of 46 calves kept under poor hygienic, management and preventive measures in company with other animals of different ages in a dairy farm were suffering clinically from enteritis manifested by diarrhea. These calves were used for collection of rectal swaps for isolation and identification of E. coli. Diarrheic calves were examined for signs of enteritis with detection of fecal consistency, odor, color and presence or absence of mucus and blood.
Reference Controls for E. coli Strains
E. coli Standard serotype O101 was obtained from Aerobic Bacterial Vaccine Department, Veterinary Serum and Vaccine Research Institute, Abbasia, Cairo, Egypt. The standard strain was subjected to bacteriological identification and the DNA was extracted for using in PCR as positive controls while PCR grade water was used as a negative control.
Bacteriological examination of fecal (Quinn et al., 2002): Fecal swabs were inoculated into trypticase soya broth and incubated at 37 °C for 24 hours for propagation of E. coli. Subcultures from trypticase soya broth were streaked on MacConkey and EMB agar media and incubated at 37 °C for 24 hours. Identification of the isolated bacteria was done based on colony morphology, staining characters and biochemical reactions.
Biochemical identification was carried out according to Quinn et al. (2002). One colony from each pure culture was picked up and streaked on TSI agar and Simmon’s citrate agar media and incubated at 37 °C for 24 hrs. After complete identification, the bacterial isolates were stored at -20 °C on brain heart infusion broth containing 16% glycerol for long term preservation.
QIAamp® DNA Mini Kit (Cat. No. 51304 Qiagen)
The QIAamp DNA Mini Kit was used for used for nucleic acid extraction.
Oligonucleotide Primers Used in cPCR
Different sets of primers were synthesized by Metabion (Germany) and supplied in a lyophilized form, reconstituted in PCR grade water to reach a concentration of 100 pmol/ μl stock solution for each primer.
Extraction of DNA was carried out according to QIAamp DNA mini kit instructions. Preparation of PCR Master Mix was carried out according to Emerald Amp GT PCR mastermix (Takara) Code No. RR310Akit.
Agarose gel electrophoreses was carried out according to Sambrook et al. (1989). The gel was photographed by a gel documentation system and the data was analyzed through computer software.
Table 1: Primers used for molecular identification of E. coli and detection of virulence factors.
| Target gene | Primers sequences | Amplified segment (bp) | References |
| ACACTGGATGATCTCAGTGG | 614 |
|
|
| CTGAATCCCCCTCCATTATG | |||
| CCATGACAACGGACAGCAGTT | 779 | ||
| CCTGTCAACTGAGCAGCACTTTG | |||
|
|
CGATTCTGGAAATGGCAAAAG | ||
| CGTGATCAGCGGTGACTATGAC | |||
| ATGCTTAGTGCTGGTTTAGG | |||
| GCCTTCATCATTTCGCTTTC | |||
| AACAAGGATAAGCACTGTTCTGGCT | |||
| ACCATATAAGCGGTCATTCCCGTCA | |||
| GAAACAACATGACGGGAGGT | 229 | ||
| GCACAGGCAGGATTACAACA | |||
| GGTTTCTGCGTTAGGTGGAA | 606 | ||
| GGGACTTCGACCTGAAATGT |
Results
A total of 14 (30.43%) calves showed signs suggestive for enteritis. Clinical abnormalities included fever, diarrhea, dehydration, recumbence and death of two calves. It was observed that these calves were unable to suckle their dams effectively delaying taking of colostrum from their dams at the proper time and predisposing them to infection.
The results of bacteriological examination of 14 rectal swaps collected from diarrheic calves revealed isolation of 13 (92.85%) suspected E. coli isolates that were inoculated onto TSI agar and simmon’s citrate agar media. On MacConkey agar, E. coli appeared as flat and dry colonies of 2-3 mm in diameters with pink color due to fermentation of lactose and surrounded by dark pink area of precipitated bile salts. On EMB agar, E. coli appeared as flat colonies of 2-3 mm in diameter and dark violet in color with a black center which produces a distinctive green metallic sheen when light was reflected on it.
The results of biochemical identification proved the presence of E. coli. E. coli fermented all sugar and produced yellow slant and butt with production of gas but did not produce H2S. Citrate utilization showed negative reaction, Vogues Proskauer and Urease tests revealed negative reaction, while Indole and Methyl red tests showed positive reaction confirming E. coli identity.
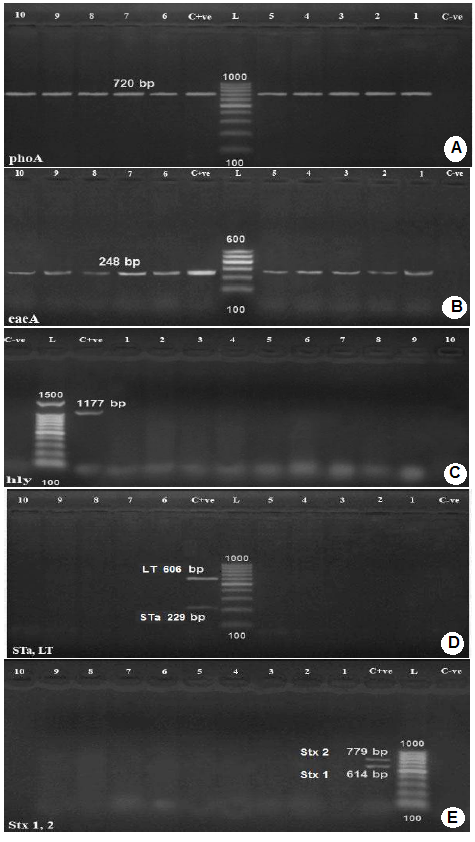
Amplification of phoA gene specific for E. coli using PCR (Figure 1-A) revealed that, all the 13 E. coli isolates showed positive result for the presence of phoA gene.
Genotyping of E. coli isolates was carried out using Multiplex PCR for the amplification and detection of the genes
Lanes 1,2,3,4,5,8,9,10,11,12: positive samples with band of amplicon size at 720 bp.
C+ve: Positive control.
L: Molecular weight marker (100 bp.)
C-ve: Negative control.
B): Agarose gel electrophoresis of PCR for detection of eaeA gene in E. coli isolates.
Lanes 1-10: positive samples with band of amplicon size at 248 bp.;
C+ve: Positive control.
L: Molecular weight marker ( 100bp.)
C-ve: Negative control.
C): Agarose gel electrophoresis of PCR for detection of hly gene in E. coli isolates.
C-ve: Negative control.;
L: Molecular weight marker (100 bp.)
C+ve: Positive control.
Lanes 4-13: negative samples.
D): Agarose gel electrophoresis of PCR for detection of Sta and LT genes in E. coli isolates.
Lanes 1,2,3,4,5,6,9,10,11,12: negative samples
C+ve: Positive control.
L: Molecular weight marker (100bp.);
C-ve: Negative control.
E): Agarose gel electrophoresis of PCR for detection of stx1 and stx2 genes in E. coli isolates.
C+ve: Positive control.
L: Molecular weight marker (100bp.);
C-ve: Negative control.;
encoding shiga toxin1(stx1), shiga toxin 2 (stx2), intimin (eae), hemolysin (hylA) and E. coli enterotoxin genes such as heat stable enterotoxin (st), and heat labile enterotoxin (lt). All tested E. coli isolates were positive for intimin (eae)A, attaching and effacing gene (gene species specific) for E. coli (100%). No isolate had shiga toxin1 (stx1), shiga toxin 2 (stx2), hemolysin (hylA), and E. coli enterotoxin genes heat stable enterotoxin (st) and heat labile enterotoxin (lt), Figure 1 (B,C,D,E).
Discussion
Clinical abnormalities suggesting enteritis were recorded in 14 (30.43%) calves of different ages (1-4 weeks) that showed fever, diarrhea, dehydration, pneumonia recumbence and death of two calves. Similar clinical picture was reported by Al-Alo et al. (2018), Cho et al. (2010) and Singh et al. (2013). Hossain et al. (2013) reported that the symptoms of calf enteritis included diarrhea, a rise in body temperature, general weakness, dehydration and lack of appetite followed by coma and death within a few hours. The high morbidity 30.43% recorded in this study among new born calves may be attributed to several factors including the calf less mature immune system than in adults, agammaglobulinemia and the high level of hydrocortisone that probably due to transplacental transfer, as well as the fetus produces large quantity of corticosteroids beginning 8–10 days before birth that lead to immunosuppression as discussed by Hosein (2018).
Identification of 13 (92.85 %) isolates of E. coli out of the 14 collected samples of diarrheic calves was confirmed on the basis of their morphology, cultural and biochemical tests using standard bacteriological procedures. The 13 isolates were visualized as blue-black colonies (2-3mm diameter) with green metallic sheen on Eosin Methylene Blue Agar and bright pink colonies on MacConkey agar. Such findings indicated a presumptive identification of E. coli that was confirmed using standard bacteriological procedures.
The overall prevalence rate of colibacillosis in the present study among 46 calves was (28.26 %). This may be regarded to the fact that E. coli is a normal inhabitant in the intestine of warm-blooded animals as mentioned by Achá et al. (2004). The present result agreed and disagreed in respect of the prevalence of infection with the findings of different authors; Bashir et al. (2015), (35%); Cookson et al. (2006), (88.7%); Pourtaghi et al. (2013), (86.7%) and Shahrani et al. (2014), (76.45%). The difference in the rates of E. coli infection may be attributed to differences in preventive and hygienic measures and exposure to stress factors allowing the opportunistic E. coli to flourish and express its virulence genes causing pathogenic effect on calves as mentioned by Cho and Yoon (2014).
A monoplex PCR detecting alkaline phosphatase gene (phoAgene) specific for E. coli was performed on E. coli isolates. The results of the amplification of phoA gene using PCR, (Figure 1) revealed that all E. coli isolates showed positive result for the presence of phoAgene, thus confirming their identity as E. coli and proving that phoA gene is a housekeeping gene present in all E. coli strains as reported by Chang et al. (1986) and Kong et al. (1999). In order to categorize the different pathotypes of pathogenic E. coli, multiplex PCR was used for detection of virulence genes encoding virulence factors including shiga toxin1 (stx1), shiga toxin 2 (stx2), intimin (eae), hemolysin (hylA), and E. coli enterotoxin genes such as heat stable enterotoxin (st) and heat labile enterotoxin (lt). All tested E. coli isolates were positive for intimin (eae) A, attaching and effacing gene (gene species specific) for E. coli (100%). No isolate had shiga toxin1 (stx1), shiga toxin 2 (stx2), hemolysin (hylA), and E. coli enterotoxin genes heat stable enterotoxin (st) and heat labile enterotoxin (lt), photos (2-5). Intimin is a virulence factor of EPEC (e.g. E. coli O127:H6) and EHEC (e.g. E. coli O157:H7) E. coli strains. It is an attaching and effacing (A/E) protein, which is responsible for enteropathogenic and enterohemorrhagic diarrhea, (Stevens et al., 2006). The intimin gene detection is mainly linked to the EPEC pathotype (Beraldo et al., 2014) and eaeA-positive strains are considered to be more virulent to human than the eaeA-negative ones. This indicates a possible participation of calves in the zoonotic transmission of pathogenic E. coli as supported by the findings of Thiry et al. (2017). Nataro and Kaper (1998) reported that EPEC pathoype strains had a characteristic attaching and effacing (A/E) effect on the gut mucosa due to intimate bacterial adhesion to the enterocytes and effacement of the brush border microvilli. The formation of A/E lesions is governed by the locus of enterocyte effacement (LEE). This locus contained the eaegene, which encoded an outer protein called intimin, necessary for intimate attachment to epithelial cells. EPEC is a cause of diarrhea in ruminants and human and their pathogenicity is attributed to the production of protein intimin which was encoded by the eaegene as mentioned by Horcajo et al. (2012).
EPEC represented 100% of the tested E. coli strains. All isolates were obtained from diarrheic calves which indicated that EPEC is a calf pathogen as mentioned by Moxley and Smith (2010).
The rate of EPEC among pathogenic E. coli was investigated by many authors, Badouei et al. (2016), 1.33%, Nguyen et al. (2011), 2.6%, Arya et al. (2008), 7.7%, Orden et al. (1998), 8.1%, Wani et al. (2003), 9.73%, Islam et al. (2015) 12.5% and Güler et al. (2008) 13.3%.
In general, the high prevalence of EPEC among calves may be attributed to bad management system, lack of hygiene, overcrowding, insufficient attention to the newborn calves with insufficient intake of colostrum and lack of effective preventive measures as well as stress factors that allow the opportunistic E. coli to flourish and express virulence genes causing pathogenic effect on calves as mentioned by Cho and yoon (2014).
Conclusion
EPEC infection is a major health problem among calves and suggests the significance of poor hygiene measures in the investigated farm.
E. coli infection is multifactorial in nature and it seems to be a result of interaction between different factors that may contribute to the buildup of infection. These include the immunological, nutritional, environmental and management factors.
The results indicate the possible participation of calves in the zoonotic transmission of pathogenic E. coli.
Conflict of Interest
None of the authors has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper. All authors read and approved the final manuscript.
Authors Contribution
All authors contributed in creating this article and approved the final manuscript.
References