Advances in Animal and Veterinary Sciences
Research Article
Biosynthesis, Optimization and Characterization of Silver Nanoparticles Using a Soil Isolate of Bacillus pseudomycoides MT32 and their Antifungal Activity Against some Pathogenic Fungi
Mohamed T. El-Saadony*, Nahed A. El-Wafai, Hassan I. Abd El-Fattah, Samir A. Mahgoub
Agricultural Microbiology Department, Faculty of Agriculture, Zagazig University, Zagazig 44511, Egypt.
Abstract | Optimization of some experimental growth conditions for biosilver nanoparticles synthesized using supernatant of Bacillus pseudomycoides MT32 were studied. The optimized growth conditions were obtained at 2mM silver nitrate concentration, 30°C temperature degree, 6.5 pH level, 40 h incubation time, 140 rpm agitation speed, medium type was Nutrient broth (NB) and supernatant and silver nitrate ratio was 20:30. Characterizations of the produced nanoparticles were done using seven advanced instruments. The ultraviolet-visible spectrashowed an absorption peak at 420 nm. Transmission Electron Microscopy (TEM) showed that the mean diameter of the formed SNPs was 25 to 43 nm. Powder X-ray diffraction (XRD) revealed that the particles are crystalline in nature, with a spherical structure and their size ranges from 32 to 86 nm. From dynamic light scattering (DLS) and Zeta potential analyses, the average SNPs size was 63.39 nm and the Zeta potential was −18.3 mV. Energy-dispersive X-ray spectroscopy (EDX) exhibited strong signal in the silver region and confirmed the formation of SNPs. The SNPs also exhibited traces of agglomeration. From antifungal studies in vitro, the produced SNPs are capable of suppression, in different extents, of ten commercially plant pathogenic fungi and the recorded values of MIC and MFC were varied due to the type of fungi used, and they were in a range of 70 – 90, and 75 – 100 µg / l, respectively. The antifungal activity of the SNPs produced by Bacillus pseudomycoides MT32 is useful in solving different problems in crops production as well as in animals’ nutrition.
Keywords | Optimization, Characterization, Bacillus pseudomycoides MT32, SNPs, Antifungal activity
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | November 18, 2018; Accepted | December 03, 2018; Published | January 18, 2019
*Correspondence | Mohamed T. El-Saadony, Agricultural Microbiology Department, Faculty of Agriculture, Zagazig University, Zagazig 44511, Egypt; Email: m_tlatelsadony@yahoo.com
Citation | El-Saadony MT, El-Wafai NA, El-Fattah HIA, Mahgoub SA (2019). Biosynthesis, optimization and characterization of silver nanoparticles using a soil isolate of Bacillus pseudomycoides MT32 and their antifungal activity against some pathogenic fungi. Adv. Anim. Vet. Sci. 7(4): 238-249.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.4.238.249
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 El-Saadony et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
In fact, the biosynthesis of nanoparticles has received considerable attention in the last decades due to their intrinsic merits i.e., ultrafine size, high surface area to volume ratio, optical, electrical, magnetic, catalytic and thermal properties (Chowdhury et al., 2016). Silver nanoparticles (SNPs) is defined as a silver mineral with very small size (10-100 nm). They attracted the attention of workers in different fields all over the world due to the unique chemical and physical properties and they are generally having today a promising application in medicine, agriculture, environment remediation, food technology, water treatment etc. (Singh et al., 2015). Different microorganisms i.e. bacteria, actinomycetes, and fungi have been studied for synthesis of bio silver nanoparticles. They synthesized SNPs when grab target ions (Ag+) from their habitats and turn metal ions into the elemental form (Ag0) through generated enzymes, NADH-and NADPH- dependent reductase enzyme (Benzerara et al., 2010).
In recent years, plenty numbers of studies are focused mainly on microorganisms due to the fact that synthesis of silver nanoparticles (SNPs) by microorganisms is more convenient than other ways and bacteria, in question are silver (S) resistant due to the specific genetic and biochemical mechanisms (Singh et al., 2015), and they are also, fast growth, easy to cultivate and handle and their ability to adapt under extreme conditions (Tripathi et al., 2017). Among bacteria, earlier studies on Bacillus species have been conducted for the biosynthesis of SNPs i.e., B. lichenformis (Kalimuthu et al., 2008), B. cereus (Ganesh Babu and Gunasekaran, 2009), B. subtilis 168 (Puga-zhenthiran et al., 2009), B. stearothermophilis (El-Batal et al., 2013), B. subtilis EWF.46 (Velmurugan et al., 2014), Bacillus sp. HAI4 (Mojtabu et al., 2016), thermophilic Bacillus sp. AZ1 (Deliou and Goudarzi, 2016) and others, but the properties of stability, sizing and dispersion of the formed SNPs differed with the type of microbial species and strains used (Prakasham et al., 2012). This means that the biochemical and genetic nature of the used strain from bacteria play an important role in controlling SNPs biosynthesis, and therefore, studies searching for new microbial strains as highly efficient SNPs producers have never stopped. As for silver as a noble metal, it is nontoxic, safe inorganic antibacterial agent used for centuries, since Roman time, and has the ability of killing a lot of diseases causing microorganisms and used as antibacterial (Franci et al., 2015), antifungal (Jo et al., 2009; Kannan et al., 2015) and antiviral agents (Galdiero et al., 2011). Thus, the exploration the new bacterial strain which exhibited the ability to fabricate SNPs with certain characteristic is still at the forefront of research in Nano fields.
Keeping above information into account, this the present study has been designed to use bacterial strains namely, Bacillus pseudomycoides MT32 that was recovered from soil polluted with heavy metals for biosynthesis, optimization and characterization of SNPs produced as well as to evaluate their inhibitory action against some commercially plant pathogenic fungi, commonly found in Egyptian agriculture.
MATERIALS AND METHODS
Bacterial Isolation and Selection
Samples of soil were gathered from industrialized area polluted with heavy metals at Tenth of Rhamadan region, Sharkia Governorate, Egypt. An obtained soil was transported to the laboratory in sterile polythene bags. Sterile saline solution (0.9% w/v) was used to dilute the soil sample and the bacterial isolates were recovered by spread plate technique on nutrient agar (NA) at 30˚C for 2 days. All isolated bacteria were individually grown in 250 ml conical flasks containing 100 ml sterile nutrient broth (NB) supplemented separately either with 0.1mM AgNO3 (as primary screening) or 1.0 AgNO3 (as a secondary screening) for selecting their potential to produce SNPs. The culture flasks were put for 48h in a shaker incubator at 100 rpm and 30ºC. Observations were recorded after 48h and 24h, in the case of primary and secondary screening respectively. The turbid appearance of the culture flasks ensured the growth and the intensity of developed color was considered for selecting the target microorganism. The proceeding of the bio reduction in the turbid flasks was monitored both visual inspection or by measuring the absorbance by UV – Vis spectrophotometer EL beshehy et al. (2015).
Preparation of Bacterial Cell Free Extract
Only one selected bacterial isolate was separately inoculated in 250 ml conical flasks containing 50 ml sterile (NB). The experimental growth conditions were adjusted at pH 7 and incubation temperature 30ºC, agitation speed at 160 rpm in shaker incubator. Followed by incubation, the enriched cultures were subjected to centrifugation at 10,000 rpm (Eppendorf) for 10min. The supernatant material was separated out and utilized as crude source of NO3 reductase enzyme for the extra cellular synthesis of SNPs (Shahverdi et al., 2007).
Biosynthesis of Silver Nanoparticles
For bioproduction of SNPs by the tested culture, 20 ml of supernatant derived from bacterial culture was mixed separately with 30 ml of 1mM aqueous solutions of filtered sterilized AgNO3, in 250ml conical flasks. Then, the reaction mixture flasks were placed at 160 rpm in shaker incubator at 30oC up to 24 h and allowed for reduction process. Also, a set of flasks were prepared as controls (the first one was contained supernatant without AgNO3 and the second one contained AgNO3 without supernatant) and incubated along with experimental samples (Shahverdi et al., 2007; ELbeshehy et al., 2015).
Identification Bacterial Isolate
The selected microbe was identified due to its morphological, biochemical and physiological characteristics by the procedures outlined in Bergey’s manual of systematic bacteriology (Logan and De Vos, 2009), then it was subjected to identification process by Matrix-assisted laser desorption ionization time of flight (MALDI TOF) mass spectrometry (Bille et al., 2012), as advanced and accurate tool to confirm the previously identification.
Optimization Factors Studied
To obtain maximum AgNPs production, different growth conditions and reactions for biotransformation of Ag+ ion to atom Ag0 by the tested microbe were optimized. These factors were incubation time (1, 8, 16, 24, 32 and 40h), temperature degree (20, 25, 30, 35 and 40ºC), AgNO3 concentration (1, 2, 3, 4 and 5mM), pH level (5.5, 6.0, 6.5, 7.0 and 7.5), mixing ratio of culture filtrate of the tested microbe and silver nitrate solution (1:4, 2:3, 3:2, 2.5:2.5 and 4:1) and media type (nutrient broth and Luria – Bertani broth were used for producing the culture supernatants of the tested bacteria), and these experiments were performed due to Safekordi et al. (2011); Elbesheby et al. (2015) with some modifications. In each experiment, the prepared conical flasks, 250 ml containing 50 ml of mixture reactions, were incubated in an orbital shaker at 160rpm, in light, taking into account the procedures of each parameter tested. At the end of incubation period, the progression of the reaction was monitored both visually inspection by naked eye and by measuring the absorbance maximum at 420nm in UV- Vis spectra by UV. Visible absorption spectroscopy (Laxco™, Alpha-1502 Alpha Series Spectrophotometer, 200 - 1000 nm).
Extraction and Purification of Silver Nanoparticles
For reach to the best yield of SNPs, the tested organism was grown under the optimized growth conditions using NB with pH6.5 and mixing ratio of culture filtrate and AgNO3 at 20:30 then incubated at 30ºC for 40h and agitation speed at 140rpm. Then, the SNPs were purified by three successive ultra-centrifugations at 17,000 rpm for 20 minutes at 4ºC, then the supernatant was separated out. The supernatant clear suspension was redispersed in sterile deionized water to remove the residual biomolecules. The purified solution was then dried using hot air oven at 60ºC for overnight (Nagarajan and Kuppusamy, 2013). Then 10 ml of deionized water was added to dried powder of SNPs and kept on a sonicator to prevent agglomeration of molecules and further used for characterization.
Characterization of Silver Nanoparticle
The particles of AgNPs (in reaction mixture, dried powder or annealed forms) were prepared due to the recommended procedures in each analysis and subjected to the following instruments: Ultraviolet–visible spectroscopy (Laxco™ dual beam spectrophotometer (model Laxco™, Alpha-1502 Alpha Series Spectrophotometer, 200 - 1000 nm) (Forough and Farhadi, 2010) for observing surface Plasmon resonance (SPR) absorbance peak of AgNPs, Fourier Transform - Infrared spectroscopy for FT - IR analysis (Bruker Tensor 37, Kaller Germany, due to Aguillar Mendez et al., 2011), for detection the interaction between proteins present in the supernatants used and the AgNPs and to identify the potential biomolecules in the bacterial supernatants. PAN analytic X-Ray diffractometer for XRD analysis, due to Kalabegishvili et al. (2012), to determine position, peak intensity and width of XRD. Scanning Electron Microscopy for SEM analysis using SEM Quanta 250 FEG, FEI company, the Netherlands, due to Pavani et al. (2013) and Transmission Electron Microscopy for TEM images using TEM JEOL 1010, Japan, due to Ganachari et al. (2012), they were used for detection the size, distribution and morphology (crystal structure) of the produced AgNPs. Zeta sizer analyzer (Nano Z2 Malven, Malvern Hills, UK), for Zeta potential analysis due to Dash et al. (2014) and Dynamic light Scattering – DLS analysis due to Chattopadhyay et al. (2013), for Zeta potential and the average size of the formed AgNPs determination.
Antifungal Activity Study In Vitro Inhibitory Effect of Snps Against The Tested Fungi
The inhibitory effect of the produced SNPs was evaluated against ten fungal species which were obtained from Agric. Dept. Microbiol. Fac. Agric. Zagazig Univ., Egypt as shown in Table (1). The employed method was carried out due to Elgorban et al. (2016), with some modification. Five ml of SNPs at different concentrations (i.e., 10, 30, 50, 70 and 90 µl) were prepared separately and poured into potato dextrose agar (PDA) medium prior to plating in Petri dishes. All plates were incubated at 28ºC. After 24h of incubation, mycelial disk (5mm) were carefully picked from the edge of seven – days of fungal cultures, and placed in the central of each Petri – dishes containing SNPs. PDA plates without any addition or AgNO3 were prepared as negative and positive controls, respectively. All plates were incubated at 28ºC for seven days. The radial growth of fungal mycelium was recorded (Cm).
Measurement of Minimum Inhibitory Concentration (Mic)
The activity of the produced SNPs at four different concentrations (i.e., 65, 75, 85and 95 µg/l) was determined against the tested fungi by broth macro dilution method due to Alizadeh et al. (2014) with some modifications. The prepared SNPs dilutions (0.5ml volumes) were prepared in glass test tubes, then 0.5 ml of the of the tested fungi with turbidity of 2.5 × 103 CFU /ml was added to each test tube. All tubes of all tested concentrations were incubated at 28ºC for 28h, and MIC was recorded as the lowest concentration of the tested agent used in this experiment resulting in the maintenance or reduction of the inoculum.
Measurement of Minimum Fungicidal Concentration (Mfc)
MFC was determined by inoculating the contents of all the prepared fungi combined with the tested SNPs concentrations in test tubes, as prepared in case of MIC test on a new PDA plates. All plates were incubated at 28ºC for 48 to 72h. The lowest concentration of the used agent resulting in no growth (Alizadeh et al., 2014).
RESULTS AND DISCUSSION
Screening and Identification of Used Bacterium
Initial, only 3 bacterial isolates out of the total 29 isolates collected from the polluted soil sample, when the first concentration (0.1 mM AgNO3) was used (first screening), were chosen. On screening these isolates using the second concentration (1mM AgNO3 in the second screening) only one bacterium out of 3 was selected for further studies. This isolate was coded MT32 and it was selected due to its ability and the intensity to change the color of reaction mixtures at the two tested concentrations and two tested incubation times (48 and 24h). The pure isolate was Gram positive, nonmotile, long rod and spore forming under light microscope and the results showed that this bacterium resembles and relates to the Bacillus species. From the results of morphological, biochemicals and physiological testes carried out on the selected bacterium due to procedures outlined in Bergey’s Manual (Logan and Devos, 2009) it can be inferred that this bacterium is Bacillus pseudomycoides and it was nominated as Bacillus pseudomycoides MT32. This bacterium was further subjected to rapid and accurate identification of bacteria and fungi by MALDI TOF Mass spectrometry.The obtained result showed its maximum identity of 98% to various Bacillus spp. mainly Bacillus pseudomycoides DSM 12442T DSM. (Krasny et al., 2013). Thus, the local bacterial isolate, Bacillus pseudomycoides MT32 is similar Bacillus pseudomycoides DSM 12442T DSM.
Biosynthesis of Snps By the Tested Microorganism
Experimental growth conditions had a strong effect on the properties, size, morphology and stability of the metal NPs. A major field of interest is how to create a biosynthesis method to control the morphology, properties, stability and size. The progression of the reaction mixture in the turbid flasks was initially observed by color change. The difference in color of reaction mix from shallow yellow to brown within a day of inoculation indicated the biosynthesis of SNPs by bacterial extract of the selected bacterium Figure (1) in the tested mixture. Control treatments displays no color change when incubated at the same conditions. It is well known that excitation of surface Plasmon trembling of metal NPs show yellowish brown color in water (Kalaiselva, 2013). An excitation of surface plasmon trembling which is distinguishing of silver nanoparticles causing changing in the color (El-Raheem et al., 2011). An electromagnetic field in the observable variety is united to the collective oscillation of transmission electrons, the dipole oscillation arising the surface Plasmon vibrations will happen (Mubayi, 2012).
Optimization Factors Used
The biofabrication of SNPs by Bacillus pseudomycoides MT32 was optimized through seven growth condition and reaction, using a primary temperature degree at 30C, incubation time at 24h and agitation speed at 160rpm as an appropriate value commonly used.
Mixing Ratio of Culture Supernatant And Agno3
After an incubation of 5 different volumes of 1mM AgNO3 with 5 different volumes of the obtained supernatants of B.pseudomycoides MT32 (10:40, 20:30, 25:25, 30:20 and 40:10) for 24h at 30ºC, UV- visible spectra of SNPs were determined, and are depicted in Figure (2). UV - Vis readings obtained in this experiment showed the absorbance of the reaction mixture were 0.9, 1.0, 0.8, 0.6 and 0.5 at 420 nm for the tested ratios, respectively. These results showed that volumes of 1mM AgNO3 which was mixed with supernatant at 20: 30 and was considered to be the optimal ratio since it gave a maximum absorbance of 1.0 at 420nm. The increase in culture filtrate volumes causes a decrease in absorbance. This reduction recommends reduction in NPs size. This means that by increase of the amount of filtrate in the mixture the amount and size of nanoparticles become smaller (Safekordi et al., 2011).
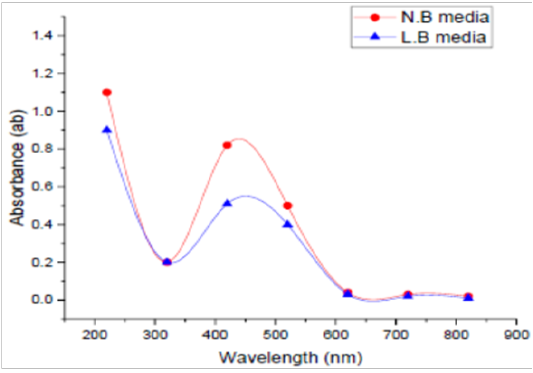
Medium Type
Two different media, LBB and NB, were used to detect the best one in this experiment. UV Vis spectra of SNPs by the tested microorganism in LBB and/or NB are presented in Figure (3). The absorbance of LBB broth was 0.61, whereas this absorbance in NB was 0.77 at 420 nm. NB was found to be the optimized medium for the manufacturing of SNPs by the tested microorganism. As for medium type, Koilparambili et al. (2016) reported that NB was found to be the standard media for the production of SNPs by Escherichia coli.
Temperature Degree
Five different temperature degrees were tested in this experiment i.e., 20, 25, 30, 35, and 40ºC. It was easy to notice that 30º C was found to be the standard temperature for the manufacture of SNPs by the tested organism (Figure 4). Also, the result showed that increase in temperature decrease the bio formation of SNPs and this reduction could be due to the inactivation or degradation of biomolecules responsible for the bio reduction process. Reaction rate increases causing consuming of reaction temperature increases most silver ions to be in the formation of nuclei and consequently the secondary reduction procedure on the surface of nuclei preformation has been stopped (Safekordi et al., 2011). As well, the study of Amit et al. (2012) reported that absorbance increased with increase in the temperature from 25 to 45°C and thereafter decreased at higher temperatures.
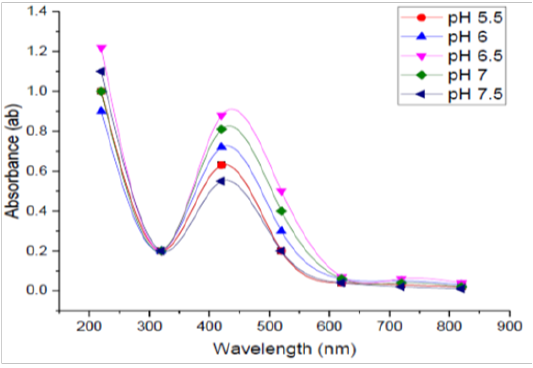
pH Levels
Five different pH values namely i.e., 5.5, 6, 6.5, 7, and 7.5 were selected for optimization process. UV - Vis readings were obtained after incubation and are shown in Figure (5). It was found that pH 6.5 was recorded to be optimum for the manufacture of silver nanoparticles by B. pseudomycoides MT32. Many studies have reported an increase in manufacture of SNPs at lower pH and the absorbance was high record was observed at this pH level. The NO3- reductase enzyme catalyzing the biosynthesis of SNPs is probably deactivated as a condition became more alkaline and this may be the reason why reduced synthesis and lower absorbance were noticed at higher pH values (Koilparambili et al., 2016).
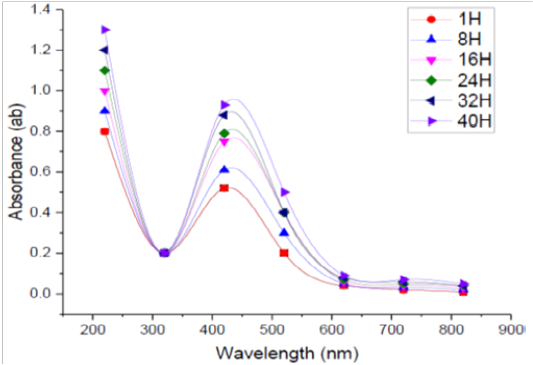
Incubation Time
The absorption spectra of SNPs formed in the tested medium at different durations every 8 h. were studied. Figure (6) shows the UV-Vis spectra of bioreduction of Ag to SNPs using supernatant prepared from the tested bacilli at different reaction times. It was found that when the incubation time was less than 1h. there was no formation of SNPs because the redox potential of the silver nitrate is reduced. Moreover, it was observed that by increasing the time of reaction, the absorption peak was increased, and more SNPs were formed during the studied incubation time (1–40 h.). From the recorded spectra, it was found that the optimum incubation time to complete the reaction in this experiment was 40 h, were a maximum absorbance was recorded otherwise, the incubation time was over. In general, the incubation needed for NPs fabrication by microorganism’s ranges from 1 to 5 days, and the properties of the size, stability and dispersion varies due to the type of microbial strain used (Prakasham et al., 2012). In this respect El-Batal et al. (2013) reported that the maximum SNPs synthesis by Bacillus stearothermophilus after 72 h. The intensity of the color of the reaction mixture was found directly proportional to the incubation time (Ibrahim, 2015).
Agno3 Concentration
Different concentrations of AgNO3 solutions (1, 2, 3, 4 and 5mM) were conducted to maximize the yield of SNPs. As can be seen in Figure (7), the UV-Vis. absorption spectra showed that by increasing the concentration of AgNO3 solution and its absorbance of SNPs was amplified, and the maximum yield of SNPs was obtained when the concentration of AgNO3 solution was 2mM, where the maximum absorbance was recorded at 420. As for the optimal concentration. Thamilselvi and Radha (2013) reported that Pseudomonas putida NCIM 2650 synthesized maximum silver nanoparticles with 1mM AgNO3. Also, Chowdhury et al. (2016) came to the same concentration. Whereas, Amit et al. (2012) found that there was a noticeable increase in yield of SNPs as the metal salt concentrations increased from 0.5 to 4mM.
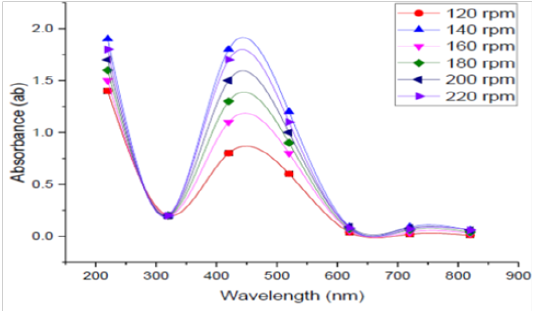
Agitation Speed
Effect of agitation speedon bioproduction of SNPs by the tested bacteria was detected at different agitation conditions 120, 140, 160, 180, 200 and 220 rpm in shaking conditions, and the obtained reading are depicted in Figure (8). The optimum rpm to biosynthesis silver nanoparticles by B. pseudomycoides MT32 was 140rpm in this experiment.This finding agrees with the results of Song et al. (2012), since they stated that the size of SNPs decreased when agitation speed increased at 250 rpm. Higher agitation speed provided a more homogenous suspension environment and hence increased the reaction surface area.
Characterization of the Produced Silver Nanoparticle Visual Inspection
It was found that the greenish yellow color appearance in the solution and mirror similar lighting on the Erlenmeyer flask walls obviously pointed out the biofabrication of SNPs in the interaction mix and the reduction of Ag+ ions to elemental SNPs (Ag0) was approved by the used bacterium Figure (9) The change in color shows the creation of SNPs.
Uv-Vis Spectra
Surface Plasmon band centered at nearly 420 nm which is showed by using the UV-Vis spectra of the SNPs (Figure 10), which is the distinguishing of SNPs and visibly pointed out the synthesis of SNPs in reaction mixture. The wavelength, at which the imaginary and real portions of the dielectric function of silver practically disappeared was ~320 nm. The Plasmon bands are wide with an absorption tail in the longer wavelengths, which could be in practice because of the particle allocation size. The accurate attitude of absorbance relies on several elements like the medium dielectric constant, size of the particle, etc. In the present study, chemical reduction process was accompanied for the manufacture of SNPs and the SNPs presented an absorption peak at 420 nm. This is consistent with previous reports showing that the range of UV-V is spectra of the Ag+ colloid nearly from 200-700 nm. Sileikaite et al. (2006) reported that the absorption band in noticeable zone and plasmon peak at 445 nm is idealistic for SNPs synthesis. Similarity, Senapati (2005) indicated that a strong surface plasmon reverberation placed at 415-420 nm was exposed when the SNPs manufactured by Fusarium oxysporum strain display.

Figure 10: UV-Vis spectrum of SNPs biofabrication
Fourier Transform - Infrared (Ft- Ir) Analysis
FT-IR measurements were performed in this study to identify the potential biomolecules in the reaction mixture. The FTIR spectra of biofabrication of SNPs presented six distinct peaks, measuring 3429.14, 1650.12, 1552.67, 1387.95, 1162.98 and 618.34 cm−1 The peak at 3429.14 cm−1 mentions to O–H stretch, H–bonded vibration of alcohols, phenols. The tip of the curve at 1650.12cm−1 assignes to N–H bend vibration of primary amines. The crest of the curve at 1552.67cm−1 refers to N–O asymmetric stretch vibration of nitro compounds. The peak at 1387.95cm−1 mentions to C–H rock vibration of alkanes. The peak at 1162.98 predicted to C–O stretch vibration of alcohols, carboxylic acids, esters, ethers (Figure 11). The peak at 618.34 cm−1 refers to –C≡ C–H: C–H bend vibrations of Alkynes, Thamilselvi and Radha (2013) reported that FTIR spectra of SNPs and observed absorption peaks located in the region of 4000 cm-1 to 400 cm-1. The carbonyl groups of the amino acid residues and the peptides have strong facility to bind to the Ag (Balaji et al., 2009). The overall conclusion, hence confirmed the present of protein in the supernatant supporting the previous studies. Many researchers stated that the proteins can bind to SNPs, either through free cysteine or amine groups in proteins. These proteins which existing over the SNPs surface might acts as capping agent for stabilization. Thus, based on this spectrum fingerprint it can be inferred that many functions groups are presented and involved in the conversion of silver ions to SNPs.
Powder X-Ray Diffraction
XRD is commonly employed to explore the characteristic and structural details of the formed nanoparticles. The XRD pattern was analyzed to determine position, peak intensity and width. Figure 12. shows the XRD patterns of vacuum-dried SNPs synthesized using B.pseudomycoides MT32.The XRD patterns indicated that the structure of SNPs produced was spherical in shape in addition, all the SNPs had a similar diffraction profile, where the scattering XRD spectrum of SNPs showed four peaks at Braggs angles which confirmed to metallic silver phase, and XRD peaks at 2θ of 32.23°, 46.13°, 56.19°, and 86.27° could be return it to the 111, 200, 220, and 311 crystallographic planes of the spherical shape of silver crystals This indicates the biosynthesized SNPs are well crystallized. The obtained results are similar to those of previous reports of characterization of SNPs by XRD, (Ahmad et al., 2009; Litvin et al., 2012).The main crystalline phase was silver, and there were no obvious other phases as impurities were found in the XRD patterns (Figure 12). Pandey et al. (2012) stated that these typical XRD peak occurs due to the presence of Face centered cubic (FCC) of the crystalline SNPs. The high intensity of the recorded peak for FCC materials is generally (111) reflection, which is observed in all the samples. The XRD analysis showed that SNPs produced were crystalline in their nature. These results are in correlation with the reports of Jeevan et al. (2012) and Manivasagan et al. (2013). From data, the average crystallite size of four different size of SNPs was estimated and was found in the range of 30 to 70 nm. The overall results of XRD was correlated with the results of Theivasanthi and Alagar (2012).
Scanning Electron Microscopy (Sem).
The SEM micrograph of SNPs synthesized by the tested bacterium is presented in Figure (13). SEM presented more insight to seeking the size and morphology details of the SNPs. From the results, the size of SEM was ranged from 28.09 to 39.33 nm, Individual SNPs and some assembled spherical particles can be seen as shown in the same figure. In general, the characteristics of monodispersity, shape, size of particles were highly dependent on the function of SNPs. The obtained data from SEM analysis were very similar to the findings of other researchers (Yousefzadi and Shin, 2016).
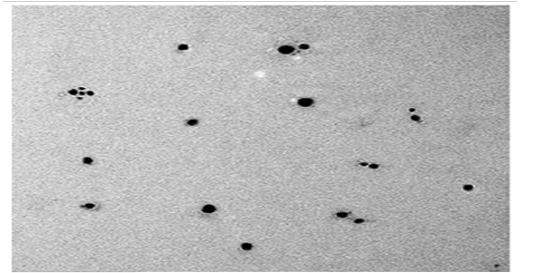
Transmission Electron Microscopy (Tem).
The TEM micrograph of SNPs produced by B.pseudomycoides MT32 is presented in Figure (14). This analysis is used to visualize size and shape of biosynthesized SNPs.It was exposed that the particle was spherical and dispersed well without agglomeration. The particle size of SNPs was sized from 25 to 43 nm. This picture shows some individual particles. In this respect, Rathod et al. (2016) stated the biosynthesis of spherical SNPs within the size range of 5– 50 nm by Nocardiopsis valliformis, and Pseudomonas mandelii synthesized the least sized SNPs with an average diameter of 1.9–10 nm. On contrary, analysis of FESEM demonstrated that SNPs were produced by Bacillus sp. HAI4., as spherical shapes with size of 33 to 264 nm (Mojtaba et al., 2016). The worthwhile thing in nano fields, variation in nano shape and size of SNPs formed by biological system is commonly recorded due to growth and operation conditions as observed from the practical experience.
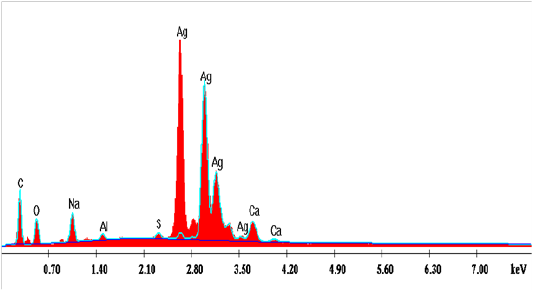
Edx Spectroscopy Analysis
The graph obtained by the EDX analysis showed the attendance of the SNPs (Figure 15). The EDX investigation assured that the existence of SNPs in the reaction mixture, giving a characteristic peak at 2.5–3.5 keV in EDX image, which mentions the reduction of Ag+ to Ag0. Sharpen signal of silver (S) cames from the SNPs and its atomic percentage was 47.85% also, it can be seen that some other peaks there observed. The atomic percentages of Carbon (C) 22.09%, Oxygen (O) 16.31%, Sodium (Na) 8.32%, Aluminum (AL) 1.04%, Sulfur (S) 0.71% and Calcium (Ca) 3.66%. Also, clear signal of carbon came from the adsorbed components of the microbe supernatant as well as coating material of the instrument. The signal of oxygen may be partial originated from air atmosphere or -OH group comes from the sodium hydroxideutilized in pH adjustment during the bio fabrication of SNPs. Sodium (Na) signal may be produced from the NaOH which was used for pH adjustment during fabrication of SNPs. Except (C), other elements have a very low atomic percentage compared to Ag, and suggest the fabrication of pure SNPs.The obtained results are in harmony with those reported by Magudapatty et al. (2001) since they reported that silver nanoparticles show typical absorption peak approximately at 2.5 KeV due to Surface Plasmon Resonance (SPR).
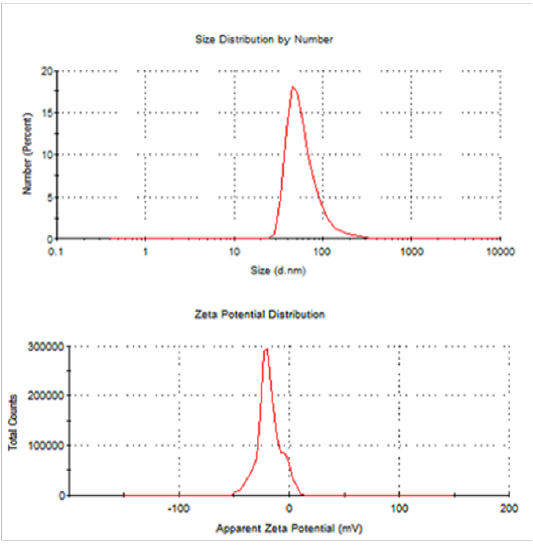
Dynamic Light Scattering (Dls) And Zeta Potential
From DLS data, it showed that the synthesized SNPs average size are 63.39 nm and 0.431 PDI value. The recorded single peak was clearly defined and this denoted that the synthesized SNPs quality is good (Figure 16) (Mahl et al., 2011). A single peak with the zeta potential of the SNPs value as -18.3mVshowed that the repulsion between the synthesized SNPs is present. A great positive or negative zeta potential of particles in suspension was recorded, this means that there will be no tendency of the particles to assemble together. Also, DLS analysis revealed that AgNPs synthesized by Bacillus pumilus, B. persicus and B. licheniformis were in the range size of 77 – 92nm, Elbeshehy et al. (2015). It is evident that the SNPs are polydispersed in nature, due to its high negative zeta potential; thus, the electrostatic repulsive force between them results in the prevention of agglomeration of the nanoparticles and is also very much helpful for long-term stability in the solution (Kotakadi et al., 2016).
Antifungal Study In vitro
The antifungal activity of the produced SNPs by B. pseudomycoides MT32 at 5 different concentrations was evaluated by three methods namely: poison plate technique, the minimum inhibitory concentration (MIC) and the minimum fungicidal concentration (MFC) using PDA medium in the presence of ten fungal species commonly found in the Egyptian agriculture and causing different problems in crops production as well as in animals nutrition, hygiene and performance, as shown in Table (1).
Inhibition Effect of Snps
Initial, in control treatments, there was a slight inhibition in the fungalgrowth of all the tested fungal species when treated with AgNO3 at 10µg / ml as a positive control when compared with PDA plates used without addition as negative control.As for the inhibition of the produced SNPs applied,as shown in Table (1), it was easy to notice that the inhibition of fungal growth, in most cases, was recorded at concentration of 90 µg / ml followed by 70 µg / ml. In addition, all the tested fungi showed a clear growth inhibition with the increases in the concentration of SNPs applied, but this inhibition was varied due to the used SNPs concentration as well as nature of the used fungi.Thus, the obtained results suggested that the produced SNPs byB. pseudomycoides MT32 are capable of suppression of the tested pathogenic fungi, in the meantime, results vary due to the concentrations of SNPs applied and the type of fungi used.The obtained results are in harmony with those reported previously (Kim et al., 2012; Alizadeh et al., 2014; Barzegar et al., 2018), since the latter investigated the effect of antifungal activity of synthesized nanoparticles, methanolic extract of Zygophyllum qatarense Hadidi leaf and silver nitrate against Aspergillus niger and Penicillium digitation by disk diffusion and micro broth dilution methods. They reported that all of the treatments showed antifungal activity, but silver nanoparticles when compared with other treatments had a significant effect against the Aspergillus niger and Penicillium digitation. In general, many studies have stated that the effect of SNPs on bacteria is strongly influenced by their size, shape and concentration (Franci et al., 2015).
With regard to the results of MIC of the tested fungi, where four concentrations of SNPs were employed namely 65, 75, 85 and 95 µg/ml, in potato dextrose (PD) medium, it was easy to infer that the lowest inhibition was recorded against the fungus Penicillum notatum being 70 µg/ml. Also, the highest inhibition of fungal growth was noticed against Rhizoctonia solani and Trichoderma viride being 95 µg/ml, in PD medium.
On the other hand, the lowest MFC was recorded 75 µg/ml in the case of Penicillum notatum and the highest MFC was noticed in the case of Rhizoctonia solani being 100 µg/ml followed by 95 µg/ml in the case of Fusarium oxysporium, where the lowest concentration of SNPs as antifungal
Table 1: Inhibitory effect of the produced SNPs against the tested fungi
|
NO |
Test pathogen |
*Growth diameter (Cm) |
||||||||
|
*PDA Only |
**PDA+ AgNO3 |
Concentration of SNPs (µg/ ml) |
||||||||
| 10 | 30 | 50 | 70 | 90 | MIC | MFC | ||||
| 1 | Aspergillus flavus | 9 | 8 | 7 | 5 | 4 | 2 | 0 | 85 |
90 |
| 2 | Aspergillus niger | 9 |
8 |
7 | 5 | 3 | 1 | 0 | 80 |
85 |
| 3 | Aspergillus tereus | 8 | 7 | 6 | 5 | 3 | 1 | 0 | 80 |
85 |
| 4 | Penicillum notatum | 8 | 7 | 6 | 4 | 2 | 0 | 0 | 70 |
75 |
| 5 | Rhizoctonias olina | 9 | 8 | 7 | 6 | 4 | 2 | 1 | 95 |
100 |
| 6 | Fusarium solani | 9 | 8 | 8 | 6 | 5 | 2 | 0 | 90 |
95 |
| 7 | Fusarium oxysporum | 9 | 8 | 7 | 5 | 3 | 1 | 0 | 75 |
80 |
| 8 | 9 | 8 | 8 | 7 | 5 | 2 | 1 | 95 |
100 |
|
| 9 | Verticillium dahliae | 8 | 8 | 7 | 5 | 3 | 2 | 0 | 75 |
80 |
| 10 | Pythium spinosum | 9 | 8 | 8 | 6 | 3 | 1 | 0 | 75 |
80 |
* PDA medium was used without any addition as negative control
** AgNO3 (10µg/ ml) was used as positive control
agent resulting no growth This could be due to the high density at which the used suspension was able to saturate and cohere to fungal hypha and to deactivate plant pathogenic fungi. Thus, the obtained results support the use of SNPs at defined concentration to control these plant pathogenic fungi under the Egyptian conditions after carry out further investigations under field conditions, and previous studies also confirmed that it corresponded to this study (Min et al., 2009; Elgorban et al., 2016). Different reports on the inhibitory actions of SNPs on microorganisms have revealed that upon treatment with silver ions, DNA loss its ability to replicate, (Feng et al., 2000), resulting in inactivated expression of ribosomal, subunit proteins, as well as certain cellular proteins and enzymes essential to ATP production (Kim et al., 2012; Sang et al., 2012). As for MIC range, the MIC of SNPs produced by B. pseudomycoides MT32 was found to be in a range from 70 to 90 µg/ml against all the tested commercially plant pathogenic fungi. With regard to the difference between the recorded MIC range in this study and those reported by Akhtar et al. (2016) who reported that the MIC value of the synthesized nanoparticles lies in the range of 88 to 132 mg/ml, and their study confirmed the presence of antifungal activity of SNPs against 5 saprophytic fungi. The obtained results are found also to be more than those reported by Barzegar et al. (2018) who found that the MIC of SNPs against Penicillum digitatum and Aspergillus niger was 8 µg/ml and MFC 32 µg/ml, respectively.
CONCLUSION
From the current study it can be inferred that the soil isolate of B. pseudomycoides MT32, has proved to be a good SNPs producers and their optimized growth conditions were incubation temperature (30ºC), concentration of AgNO3 (2mM), pH of the growth medium (6.5), incubation time (40h), agitation speed (140 rpm), the ratio of culture supernatant to AgNO3 (20:30) in the presence of nutrient broth. From characterization process, the mean diameter of the formed SNPs was 25 to 43 nm and from dynamic light scattering (DLS) and Zeta potential analyses, the average SNPs size was 63.39 nm and the zeta potential was −18.3 mV. The recorded values of MIC and MFC were varied due to the type of fungi used, and they were in a range of 70 – 90 and 75 – 100 µg / l, respectively.
Acknowledgements
Authors thank their institution for support during this work.
conflict of interest
Authors declare no conflict of interests.
authors contribution
Authors were equally contributed to this work.
REFERENCES