Advances in Animal and Veterinary Sciences
Research Article
Real-Time PCRs that Discriminates Mycobacterium tuberculosis and Mycobacterium bovis Based on Single Nucleotide Polymorphism and Genome Analysis of Amplicons
V.S. Bahekar1,2*, F. Mukherjee1**, B. M. Subramanian3, S.Y. Pasha3, A. Prasad1, K.S.N.L. Surendra1, S.K. Rana4, G.K. Sharma4, D. Premalatha2, V.A. Srinivasan5
1National Dairy Development Board, R&D Laboratory, Gachibowli, Hyderabad 500032, Telangana, India; 2Jawaharlal Nehru Technological University, Hyderabad 500085, Telangana, India; 3Indian Immunological Limited, Rakshapuram, Gachibowli, Hyderabad, 500032;4National Dairy Development Board, Anand 388001, Gujarat, India;5National Dairy Development Board, 33, Telecom Nagar, Gachibowli, Hyderabad 500032, Telangana, India.
Abstract | Accurate identification of Mycobacterium strains is necessary from a diagnostic standpoint. Differences in gyrA 95 and gyrB (1410) genome due to specific single nucleotide polymorphism (SNP) were used to design Real-Time PCRs that can discriminate Mycobacterium tuberculosis (MT) from Mycobacterium bovis (MB) belonging to the Mycobacterium Tuberculosis Complex (MTBC). The Real-Time PCR employing the VIC labeled gyrA 95 probe specifically detected the ATCC reference strain of MT H37Rv and eight field strains as MT, but not MB or other members of MTBC and other micro-organism tested. The VIC labeled gyrA 95 assay could detect up to 450 fg of MT. The FAM labeled gyrB (1410) specifically detected MB, but not MT or other members of MTBC and other micro-organism tested. The assay could detect up to 600 fg of MB, but it could not detect MB from any field isolate. Both assays were completed in 78 minutes. The results of both Real-Time PCR assays did not differ significantly from culture by Student’s t-test. The analytical sensitivity of gyrA 95 assays determined by Receiver Operating Characteristic curve (ROC) was 100% at 95% Confidence Intervals (CI) |66.4 -100.0|, and analytical specificity 100% at 95% CI |75.3 -100.0|. The repeatability and reproducibility of both above assays tested by Bland-Altman Plot indicated that the mean Cq values were within acceptable statistical limits of + 1.96 SD. Comparison of partial genome sequences of gyrA 95 and gyrB (1410) of field strains of MT with MT H37Rv by Phylogenetic Tree and Disparity Index tests indicated that eight field strains were homologous to MT H37Rv, but one was homologous to Mycobacterium intracellulare. Real-Time PCR assays were found rapid, specific, sensitive, repeatable and reproducible. Further studies are necessary for conversion of these assays to quantitative formats and determine parameters of diagnostic estimates before their use under clinical settings.
Keywords | Cattle, Mycobacterium bovis, Mycobacterium tuberculosis, Mycobacterium intracellulare, SNP, Real-Time PCR, wildlife
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | July 26, 2018; Accepted | October 04, 2018; Published | November 02, 2018
*Correspondence | F Mukherjee, National Dairy Development Board, R&D Laboratory, Gachibowli, Hyderabad 500032, Telangana, India; Email: fmhyd1@gmail.com
Citation | Mukherjee F, Bahekar VS, Subramanian BM, Pasha SY, Prasad A, Surendra KSNL, Rana SK, Sharma GK, Premalatha D, Srinivasan VA (2018). Real-time PCRs that discriminates Mycobacterium tuberculosis and Mycobacterium bovis based on single nucleotide polymorphism and genome analysis of amplicons. Adv. Anim. Vet. Sci. 7(2): 49-65.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.2.49.65
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2018 Mukherjee et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Infection in domestic animals and wildlife due to Mycobacterium has been reported globally and in India (OIE, 2014; Mishra et al., 2005; Srivastava et al., 2008; Mukherjee et al., 2015). The zoonotic potential of animals infected with M. tuberculosis to human population cannot be overlooked (Srivastava et al., 2008; Mukherjee et al., 2015). In recent times the isolation of M. tuberculosis from the members of the Bovidae family has been cited, and the extent of pathogenicity caused by the identified strain has been discussed (Thakur et al., 2012; Michel et al., 2013; Malama et al., 2014; Bhanurekha et al., 2015). We had recently reported the isolation, identification and strain differentiation of M. tuberculosis field isolates from bovines and two wildlife species from India using a liquid culture medium employing BACTEC™ MGIT™ 960 Mycobacterial Detection System (Becton Dickinson India); a commercial nested PCR targeting the IS6110 sequence (Genei, Bengaluru); and reverse slot blot hybridization assays (HAIN Life Science, Germany) [Mukherjee et al.,– Manuscript accepted for publication in Revue Scientifique et Technique Vol. 37 (3)], respectively. However, in this work we wanted to explore the possibility of developing Real-Time PCR assays that could discriminate pathogenic field strains of M. tuberculosis from that of M. bovis.
Earlier, molecular assays and mini-sequencing techniques have been described using allelic variations in bacterial genomes due to existence of single nucleotide polymorphism (SNP) (Kasai et al., 2000; Gopaul et al., 2008; Bouakaze et al., 2008; Bouakaze et al., 2010). Mutations in the alleles of the members of the Mycobacterium tuberculosis complex have been used for the species and lineage specific identification, and further these mutations have been used for classification of strains broadly into three Principal Genetic Groups (PGGs) – Group 1 (a and b), 2 and 3 and SNP Cluster Groups (SCGs) (Sreevatsan et al., 1997; Brosch et al, 2002; Filliol et al., 2006; Bouakaze et al., 2010; Bouakaze et al., 2011). Thus, at the position 203 in the katG gene [represented as katG 203] the mutation ACT ACC in Mycobacterium tuberculosis strain H37Rv differentiates PGG 1a from 1b group, similarly the mutation due to SNP in sequence CTG CGG in katG 463 gene is used for assignment of strains to PGG 2 group, while presence of polymorphism in strains in ACC AGC in gyrA 95 gene indicate that is belongs to PGG 3 group (Sreevatsan et al., 1997). Earlier studies on cluster analysis of diversity in SNP nucleotides among Mycobacterium tuberculosis strains had indicated phylogenetically six distinct SNP cluster groups (SCGs) 1 to 6 and 5 sub-groups (3a, 3b, 3c, 6a and 6b) (Filliol et al., 2006). On this basis, mutations in alleles of genomes - hsp65631 CT can distinguish -M. canetti, 16S RNA 1429 TC - M. pinnipedii, gyrB (675) CT - M. microti, gyrB (765) GA - M. bovis, M. bovis BCG and M. caprae, gyrB (1410) from M. tuberculosis H37Rv (Bouakaze et al., 2010).
SNP based assays designed to discriminate strains within the genus Brucella (Gopaul et al., 2008) and Mycobacterium (Bouakaze et al., 2011) has been reported. These techniques used Real time PCR assays employing differentially labeled fluorescent probe sequences encompassing the SNP sequence to specifically discriminate closely related strains rapidly (Gopaul et al., 2008; Gopaul et al., 2010). Based on the above information and DNA sequencing data of Mycobacterium tuberculosis H37Rv available in the public domain, in this study, firstly we aimed to design probes containing the species specific SNPs and their corresponding flanking primers of selected gene targets. Secondly, by using the selected pair of primers we aimed to amplify these genes and sequence them in order to verify the presence of specific SNPs in the amplicons. Thirdly, following the affirmation of the presence of these SNPs – fluorescently labeled minor groove binding TaqMan probes targeting gyrA 95 and gyrB (1410) were employed, aimed at development of Real-Time PCRs that can discriminate between MT and MB. The check for authenticity of the assays was initially planned on reference strains, and later, their further validation on field strains. Lastly, it was aimed to compare the partial sequence information of gyrA 95 and gyrB (1410) of field and reference strains to study the degree of homology of field strain with respect reference strain of MT for epidemiological reasons.
The analysis of the authenticity of the primers and probe sequences used in this study employing in-silico analysis is described in this paper. We report here the development and validation of in-house discriminatory Real-Time PCRs targeting specific SNPs in the gyrA and gyrB genes using reference strains of Mycobacterium and field isolates from India. The significance of comparative genome analysis of partial sequences of gyrA 95 and gyrB (1410), generated from amplicons derived from DNA templates of reference and field strains of Mycobacterium is also presented in this study.
MaterialS and Methods
Sample Identification Number of Field Isolates
The identification numbers of Mycobacterium field isolates, recovered from animals belonging to the members of the Bovidae family, were given on the following basis with respect to: a) genus and species of the bacteria isolated; b) species of animal from which the isolates were recovered; c) country of origin; d) Laboratory isolation number; e) year of isolation; and f) the gene target selected for partial sequencing of amplicons from the isolates. Thus, ‘MT’ represents Mycobacterium tuberculosis; ‘MI’ -Mycobacterium intracellulare; ‘A’ – Antelope; ‘G’ – Gazelle; ‘C’ – Cattle; IG, AG, HT, MM represent the place and State of India, like GG - Gandhinagar, Gujarat, HT – Hyderabad, Telangana, MM – Mumbai, Maharashtra, respectively; IS1 to IS9 are Laboratory numbers; 2010, 2011 and 2012 represents year of isolation; and gyrA and gyrB – represents the partial sequencing of these genes from PCR products amplified from DNA extracted from field isolates. For brevity, sample identification number has been represented as IS1 to IS9 as abbreviations in the text, except as mentioned in Table 2.
Table 1: Details of primers and probes used for Normal, Real Time PCR and sequencing assays
|
Gene target/ |
Primers used for normal PCR, Real Time PCR and Sequencing |
Minor Groove Binding (MGB) differentially labelled fluorescent VIC c and FAM d probe sequence 5'3' |
Primers designed for PCR and sequencing of amplicons greater than 300 bp e |
||
|
Forward Primer (FP) and Reverse Primer (RP) sequence 53' |
Amplicon size (bp) |
Forward Primer (FP) and Reverse Primer (RP) sequence 5'3' |
Amplicon size (bp) | ||
|
16S rRNA 1429 |
FP: TGTCCAGGGCTTCACACATGC a RP: TTCACGGGGTCGAGTTGCAG a |
120 |
VIC-AAGGGCTGCGATGC FAM- AAGGGCCGCGATGC |
FP: TCATGTTGCCAGCACGTAAT RP: TTCGGGTGTTACCGACTTTC |
317 |
|
katG 463 |
FP: CGGTCGAAACTAGCTGTGAGACAGT a RP: AAGCCGAGATTGCCAGCCTTA a |
72 |
VIC- AGATCCGGGCATC FAM- AGATCCTGGCATC |
FP: ACAAGCTGATCCACCGAGAC RP: ATCTCTTCCAGGGTGCGAAT |
334 |
|
hsp65 631 |
FP: AAAAGGCCGTGGAGAAGG b RP: CAATCTGCTCCTTGGTCTCG b |
72 |
VIC -TCAAGGGCGCCA FAM –TCAAGGGTGCCA |
FP: GGTAGCCAAGAAGACCGATG RP: CCAAAGGTGTTGGACTCCTC |
321 |
|
gyrA 95 |
FP: AGACCATGGGCAACTACCAC b RP: TTTCCCTCAGCATCTCCATCb |
198 |
VIC-ACGACAGCCTGGT FAM- ACGACACCCTGGT |
FP: CAGCGCAGCTACATCGACTA RP: GGGCTTCGGTGTACCTCAT |
325 |
|
gyrB (1410) |
FP: AAGACCAAGTTGGGCAACACb RP: ACCAACTCTCGTGCCTTACGb |
167 |
VIC-CTGTAACGAACAGCT FAM- CTGTAATGAACAGCT |
FP: TGGTGAACAAGTACGCCAAG RP: ACCAACTCTCGTGCCTTACG |
301 |
|
gyrB (675) |
FP: CGATATCTGGTGGTCTGCAC b RP: GGGGTTCCGACTTCTCATAA b |
123 |
VIC- ACGGGTACGAGTGGTCT FAM- ACGGGTATGAGTGGTCT |
FP: ACGCGTATGCGATATCTGGT RP: CTCTCGTCGGTCAGGTTGAT |
301 |
|
gyrB (675+756) |
FP: AGATCAAGCGCGACGGGTAa RP: CGAAGTCGTATTCCGTGGTTTCGa |
150 |
VIC- AACGGTGCGGTTC FAM- AACGGTACCGGTTC |
ND | NS |
|
gyrB (756) |
FP: TTTCGAAACCACGGAATACGb RP: GTTCTTGGTGCGGTTGATGTb |
271 |
VIC-AACGGTGCGGTTC FAM-AACGGTACGGTTC |
ND |
NS |
aForward primer (FP) and Reverse primer (RP) - as suggested by Bouakaze et al., 2010, and b FP and RP designed In-house and used in this study for normal gel PCR, Real Time PCR and sequencing of PCR products for experimentally checking the authenticity of designed primers and the position of SNPs with respect to initial sequence homology data generated in silico derived from information in the public domain; c 2ʹ-chloro-7ʹ-phenyl-1,4-dichloro-6-carboxy fluorescein labeled (VIC) and d 6ʹ carboxy fluorescein labeled (FAM) MGB probe designed in this study for their potential use in discriminatory Real Time PCR assays; e Primers designed for amplifying DNA sequences greater than 300 base pairs from genes of Mycobacterium for comparison of partial gene sequences of field isolates from animal origin in India with reference strain of Mycobacterium tuberculosis H37Rv strain # 27294 from ATCC; ND – Not designed; NS- Not sequenced
Genomic DNA Extraction from Mycobacterium and Non-Mycobacterium Strains
The genomic DNA from field isolates of Mycobacterium (Table 2) and reference strains of Mycobacterium (Table 3) were extracted as per method described by GenoType MTBC kit from Hain Life Sciences, Germany (Cat. No. 47013). Briefly, 1 ml liquid culture from BACTEC tube was transferred to 1.5 ml micro-centrifuge tube and pelleted at 10000 xg for 15 minutes. The pellet was re-suspend ed in 100 µl of nuclease free water and bacteria were heat i-
Table 2: Details of partial sequences of gyrase A & B of Mycobacterial isolates submitted to GenBank
| Details of samples | Details of partial Nucleotide sequence(s); GenBank Accession Numbers | ||||
|
Isolate ID1 |
Source | Year of isolation | Place | Locus: gyrase A (325 nt*) | Locus: gyrase B (301 nt*) |
| MT_A_IND_IG_IS1_2010 | Antelope | 2010 |
Indroda, |
KP996515 | KP996523 |
| MT_G_IND_IG_IS2_2011 | Gazelle | 2011 |
Indroda, |
KP996516 |
KP996524 |
| MT_C_IND_GG_IS3_2010 | Cattle | 2010 | Gandhinagar, Gujarat | KP996517 | KP996525 |
| MT_C_IND_HT_IS4_2010 | Cattle | 2010 | Hyderabad, Telangana | KP996518 | KP996526 |
| MT_C_IND_HT_IS5_2010 | Cattle | 2010 | Hyderabad, Telangana | KP996519 | KP996527 |
| MT_C_IND_GG_IS6_2011 | Cattle | 2011 | Gandhinagar, Gujarat | KP996520 | KP996528 |
| MT_C_IND_MM_IS7_2012 | Cattle | 2012 | Mumbai, Maharashtra | KP996521 |
KP996529 |
| MT_C_IND_HT_IS8_2010 | Cattle | 2010 | Hyderabad, Telangana | KP996522 | KP996530 |
| MI_C_IND_HT_IS9_2010 | Cattle | 2010 | Hyderabad, Telangana |
NS 2 |
KP996531 |
1‘MT’ represents Mycobacterium tuberculosis; ‘MI’ - Mycobacterium intracellulare; ‘A’ – Antelope; ‘G’ – Gazelle, ‘C’ – Cattle; IG, AG, HT, MM represent the place and State of India, like IG – Indroda, Gujarat, GG - Gandhinagar, Gujarat, HT – Hyderabad, Telangana, MM – Mumbai, Maharashtra, respectively; IS1 to IS9 are Laboratory numbers; 2010, 2011 and 2012 represents year of isolation; and gyrA and gyrB – represents the PCR products amplified from these genes from DNA extracted from field isolates and used for partial sequencing experiments; 2 NS – Not sequenced; * nt–nucleotides.
-nactivated by incubating at 95°C for 20 minutes. Later, the bacterial cells were lysed by incubating in a water bath sonicator for 20 minutes. The lysate was centrifuged at full speed (10000 x g) for 5 minutes and the supernatant were directly used for amplification or transferred to fresh 1.5 ml micro-centrifuge tube and stored in -20°C until further use. The genomic DNA from non-mycobacterial standard strains (Listed in Table 3) were extracted as per protocol given by QiaAmp Blood Mini Kit (Qiagen, Germany). The quality of DNA was checked by agarose gel electrophoresis using 0.8% agarose.
Selection of Gene Target, Designing of Primers and Minor Groove Binding (mgb) Probes
The selection of eight gene targets (hsp65 631, katG 463, 16S rRNA 1429, gyrA 95, gyrB (675+756), and gyrB (675), gyrB (756), gyrB (1410) were based on the primary information provided in a previous publication (Bouakaze et al., 2010). However, employing the software from GenScript (www.genscript.com/tools.html#biology) new primers were designed for conducting PCR, Real Time PCR and their corresponding minor groove binding (MGB) fluorescent labeled probes and for generating amplicons longer than 300 bp for sequencing experiments for five gene targets hsp65 631, gyrA 95, gyrB (675), gyrB (756) and gyrB (1410) in this study, while retaining the original primers recommended for other three gene targets 16S rRNA 1429, katG 463 and gyrB (756+675) by Bouakaze et al. (2010); the details are provided in Table 1.
Primers designed for normal gel PCR and expected to generate amplicons less than 300 base pairs were also used for sequencing experiments. Prior to the selection of primers and probes, the potential and authenticity of sequences as candidates, for development of discriminatory assays were analyzed in silico with the help of information available in the public domain for Mycobacterium tuberculosis H37Rv strain (ATCC 27294) for Gene locus and version NC_000962.3 employing the BLASTN tool (https//www.ncbi.nlm.nih.gov>BLAST).
Normal Gel PCR and Sequencing of Amplicons
Normal Gel PCRs were performed to amplify eight gene targets encompassing the specific SNP sites which can differentiate the Mycobacterium tuberculosis (MT), Mycobacterium bovis (MB), non-MTC Mycobacterium and non-Mycobacterium species (Bouakaze et al., 2010). Primers encompassing the SNPs at different nucleotide positions within the genome were depicted as superscripts - hsp65 631, katG 463, 16S RNA1429, gyrA 95, gyrB (675+756), and gyrB (675), gyrB (756), gyrB (1410) (Table 1). PCR was performed on DNA extracts from reference strains of Mycobacterium tuberculosis procured from American Type Culture Collection (ATCC), USA strain number 27294 and Mycobacterium avium subspecies paratuberculosis from Collection de l’Institut Pasteur (CIP), Frances train number 103963, respectively, were used as templates and PCRs were performed in total reaction volume of 50 µl constituting the PCR buffer (10 X; 5 µl); dNTPs (100 mM; 1 µl);
specific primer set (10 pM; 1µl each), Taq polymerase (3U/µl; 0.8 µl) (Sigma Cat. No D6677-1.5 KU); DNA (5 µl)
Table 3: Specificity of the qPCR targeting gyrase A (gyrA) and gyrase B (gyrB) genes
| Sr. No. |
Name and origin of the straina, b,c |
Real time PCR Result | |
|
gyrA 95 gene target (Cq Value) |
gyrB (1410) gene target (Cq Value) |
||
| 1 |
Mycobacterium tuberculosis H37Rv (ATCC 27294)a |
19.28 | Negative |
| 2 |
Mycobacterium bovis BCG Pasteur (ATCC 35734)a |
Negative | 17.01 |
| 3 |
Mycobacterium avium subsp. paratuberculosis (CIP 103963)c |
Negative | Negative |
| 4 |
Mycobacterium paratuberculosis (ATCC 19698)a |
Negative | Negative |
| 5 |
Campylobacter fetus subsp. fetus (ATCC 27374)a |
Negative | Negative |
| 6 |
Campylobacter fetus subsp. venerealis (ATCC 19438)a |
Negative | Negative |
| 7 |
Brucella abortus 544 (ATCC 23448)a |
Negative | Negative |
| 8 |
Brucella abortus S19 (USDA/Mar. 98)b |
Negative | Negative |
| 9 | Saccharomyces cerevisiae (ThermoFisher Scientific Cat no. C81000) | Negative | Negative |
| 10 |
Mycobacterium phlei (ATCC 106789)a |
Negative | Negative |
| 11 |
Mycobacterium smegmatis (ATCC 607)a |
Negative | Negative |
| 12 |
Tritrichomonas fetus (ATCC 30003)a |
Negative | Negative |
| 13 | Negative control (No Template control, NTC) | Negative | Negative |
| 14 | IS-1 | 19.68 | Negative |
| 15 | IS-2 | 18.19 | Negative |
| 16 | IS-3 | 18.65 | Negative |
| 17 | IS-4 | 22.80 | Negative |
| 18 | IS-5 | 21.75 | Negative |
| 19 | IS-6 | 24.60 | Negative |
| 20 | IS-7 | 26.90 | Negative |
| 21 | IS-8 | 21.32 | Negative |
| 22 | IS-9 | Negative |
Negative |
aAmerican Type Culture Collection (ATCC); b United States Department of Agriculture (USDA); cCollection de l’ Institute Pasteur (CIP) France. IS-Field isolates of Mycobacterium.
and nuclease free water (36.2 µl). Each SNP PCR was performed for 40 cycles following the thermal profile detailed below: initial denaturation at 94°C for 5 minutes; denaturation at 94°C for 30 seconds; annealing at 60°C for 30 seconds; extension 72°C for 30 seconds and final extension at 72°C for 10 minutes. PCR products were purified using QIA Quick gel purification kit (QIAGEN, Germany) following the manufacturer’s instructions. The purified products were quantified using Nanodrop spectrophotometer and the quality and purity was checked by electrophoresis using 2% agarose gel. The eight amplicons purified as described above were subjected nucleotide sequencing by a Chain Termination method using the Big Dye Terminator Version 3.1 cycle sequencing Kit (ABI, Life Technologies, USA) by following the manufacturer’s instruction. Subsequently the labeled fragments were purified and analyzed on ABI 3130XL Genetic Analyzer (ABI, Life Technologies, USA). The primers used for sequencing were same for the corresponding gene targets as those mentioned for normal PCR in Table 1.
Development and Optimization of Real Time PCRs
In the first set of experiments, reactions were performed in 0.2 ml PCR strip-tubes (Qiagen Cat. No. 981005). The total reaction volume was 25 µL which comprises of 12.5 µL of master mix (Eurogentec qPCR master mix No ROX), 10 pico-moles of each primer (Bioserve India), 5 pico-moles of MGB probe (Invitrogen Bioservices India Pvt. Ltd) and 5 µL of the template. Serially diluted DNA, ranging from 45 ng to 4.5 ag extracted from Mycobacterium tuberculosis (MT) ATCC 27294 and 60 ng to 6 ag of Mycobacterium bovis BGC ATCC 35734 (MB). Two (gyrA 95 and gyrB (1410) out the eight gene targets (hsp65 631, katG 463, 16S rRNA 1429, gyrA 95, gyrB (675), gyrB (756), gyrB (675+756) and gyrB (1410) were tested independently in a separate series of reactions for MT and MB templates, respectively. Reaction conditions were set as follows: Hold at 50°C for 2 minutes and 95°C for 10 minutes, followed by cycling at 95°C for 15 seconds and 60°C for 60 seconds for 45 cycles. The real-time PCR assay was performed in Rotor Gene Q Real-time PCR cycler (Qiagen,Germany). The specific primers used for Real Time PCR were same as those used for normal Gel PCR (Table 1). In the second set of experiments, using the above PCR protocol, DNA extracts from MT and MB and Mycobacterium strains and from organism not belonging to genus Mycobacterium (listed in Table 3) were subjected to Real Time PCR targeting gyrA 95 and gyrB (1410) gene targets. Similarly, in the third set of experiments, gyrA 95 and gyrB (1410) targets encompassing specific SNP for MT/MB in DNA extracts of field isolates were amplified using specific primers (Table 1). For the second and third set of experiments purified DNA extracts from ATCC reference strains - MT and MB in duplicates were always included as positive control. Similarly molecular biology grade DNA free sterile distilled water in duplicates was included throughout these experiments as negative control.
Analytical Specificity and Sensitivity
Analytical specificity (ASp) of the assay was estimated by using DNA from different reference bacteria and protozoa. Analytical sensitivity (ASe) of the assay was estimated by performing the assay with known DNA concentration of standard Mycobacterium strains (MT and MB) in ten-fold serial dilution in triplicates.
Repeatability and Reproducibility
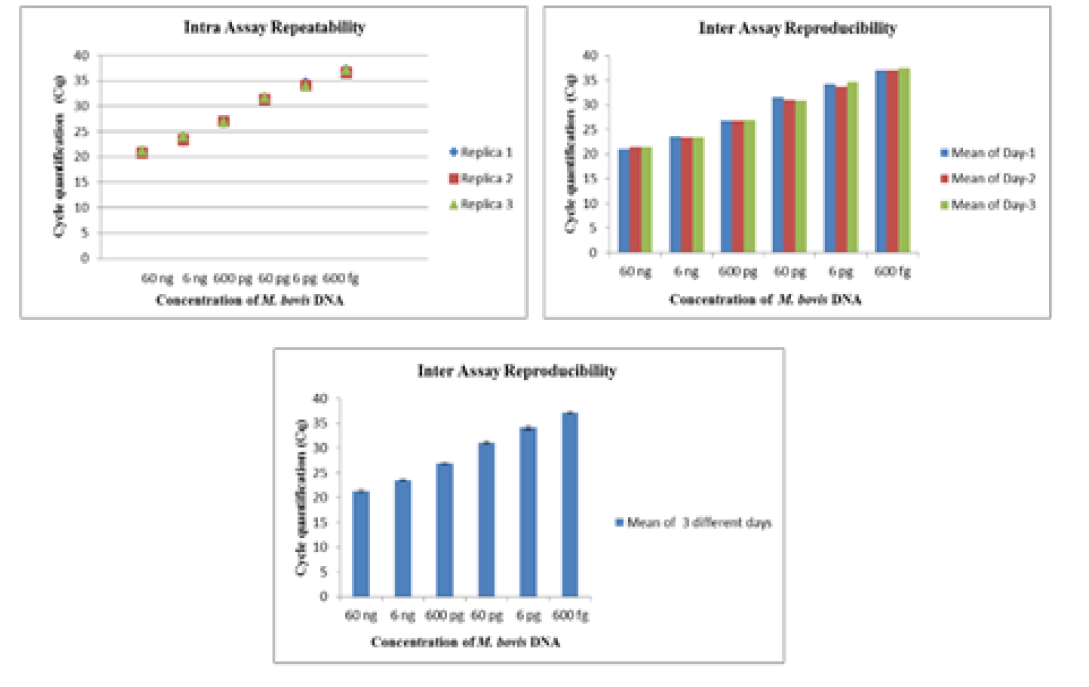
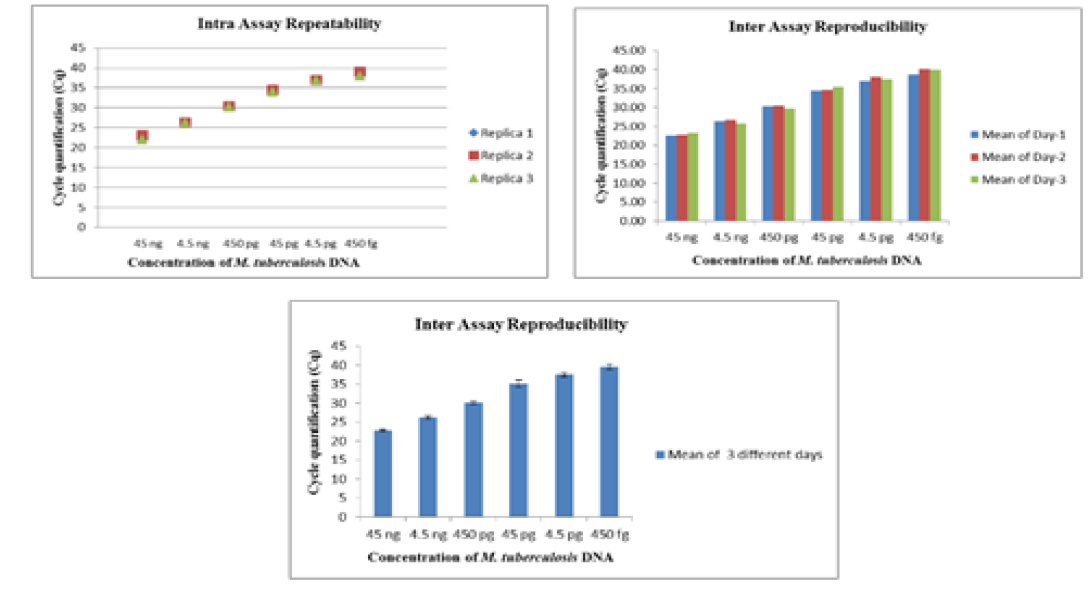
The intra-assay repeatability was determined by performing the assay using serially diluted DNA from standard strains of Mycobacterium tuberculosis (ATCC 27294) (45ng to 4.5ag) and Mycobacterium bovis BGC(ATCC 35734) (60 ng to 6ag) in triplicates on same day. The inter-assay reproducibility of the Real- Time PCR was determined by performing the assay with the same set of DNA in triplicates on three different days.
Analysis of Nucleotide Sequences
Nucleotide sequencing was performed from amplicons generated from DNA templates of field isolates which were greater than 300 bp in size, using sequencing primers (Table 1) specific for gyrA 95 and gyrB (1410). The sequences (electropherograms) obtained for each sample by paired sequencing with forward and reverse primers were analyzed using Seqscape v2.0 software. The authenticity of each base call was counter verified with the paired base call obtained in the paired sequence. Further, BLASTN analysis was performed on each sequence (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for identification of Mycobacterium species as well as to obtain the percent of nucleotide homology. Multiple sequence alignment of the partial nucleotide sequences (gyrA and gyrB) of the study along with published sequences retrieved from the GenBank was performed using Clustal W v2, in order to compare the nucleotide and aminoacid sequences. The sequences obtained were compared with the published sequence of ATCC reference strains of Mycobacterium, available in the public domain, for determining a) the actual presence of specific SNPs (Bouakaze et al., 2010), and b) degree of sequence homology of the field strain with respect to reference strains. Based on partial nucleotide sequence of gyrB (1410) from 9 Mycobacterium field isolates a Phylogenetic Tree was constructed using Maximum Likelihood Method; and sequence homology among field strains and reference strains with respect to gyrA and gyrB genes was analyzed using the Disparity Index Test.
Molecular Identification
Identification of isolates was done by using single tube nested PCR kit protocol (GeNei Amplification Reagent Set for Mycobacterium tuberculosis Complex (MTBC) group of organism, Bengaluru). The target for amplification was the IS6110 region of MTBC. Amplification using the above kit was expected to yield amplicons of size 220 bp with the outer primers and 123 bp with the nested primers. The DNA extracted from all the cultural isolates were further analyzed for identification of strains by using commercial reverse slot blot hybridization assay kits (HAIN Life Science, Germany); for differentiation of the Mycobacterium tuberculosis complex from cultured material (GenoType MTBC, Cat No 47013) was used, for additional species identification (GenoType Mycobacterium AS, Cat no 47011) was used; and finally for detection common Mycobacteria (GenoType Mycobacterium CM, Cat No 47012) was employed.
Statistical Analysis
The data on ASp and ASe of Real-Time PCRs for gyrA 95 was analyzed by using Student’s t-test and Receiver Operating Characteristic curve (ROC); but for the gyrB (1410) the Student’s t-test was used alone. The data on repeatability and reproducibility of gyrA 95 and gyrB (1410) Real-Time PCR assays were analyzed by Bland Altman Plot. For the analysis of data by Student’s t-test, ROC and Bland- Altman Plot the MedCalc® software version 14.12, 1993 – 2105 was employed.
RESULTS
PCR:
The amplified gene targets hsp65 631, katG 463, 16S rRNA 1429, gyrA 95, gyrB (675), gyrB (675+756), gyrB (756), gyrB (1410) from DNA of reference strain of Mycobacterium tuberculosis (H37Rv) were identified by their amplicon size - 72, 72, 120, 198, 123, 150, 271 and 167 base pairs, respectively (Fig 1a, 1b and Table 1). The sequencing data of the above PCR products revealed the existence SNPs at specific expected position when compared with sequence information of M. tuberculosis (H37Rv) available in the public domain, employing BLASTn analyses. Amplicons greater than 300 bp were generated from the above gene targets using sequencing primers (Table 1); specific amplification of some of the representative genes are shown in Fig 1c.
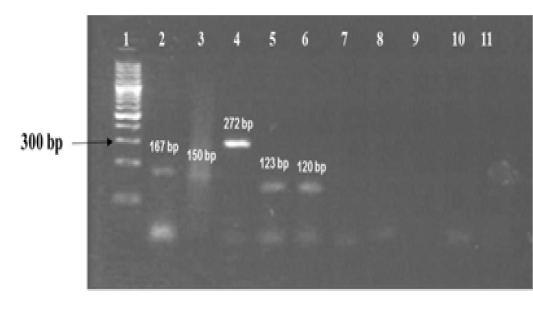
Figure 1a: Standardization of PCRs for amplification of regions encompassing SNPs for differentiation of Mycobacterium strains using DNA templates from Mycobacterium tuberculosis H37Rv (MT) strain number 27294 from American Type Culture Collection (ATCC) USA, and Mycobacterium avium subspecies paratuberculosis (MAP) strain number 103963 from Collection de l’ Institut Pasteur (CIP) France.
Lanes 1: 100 bp ladder (Fermentas, Catalogue No. SM1153); lanes 2 to 6: DNA extracts of MT ATCC strain amplified using corresponding primer sets for gyrB(1410), gyrB (675+756), gyrB (756), gyrB (675) and 16S rRNA 1429 genes respectively; lanes 7 to 10: DNA extracts of MAP amplified using corresponding primer sets for gene target gyrB (1410), gyrB (675+756), gyrB (675) and 16S rRNA 1429; lane 11: Negative control - Nuclease free water (Merck, Catalogue No. 612151181001730 )
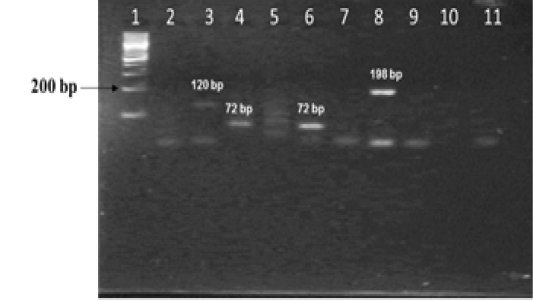
Figure 1b: Standardization of PCRs for amplification of regions encompassing SNPs for differentiation of Mycobacterium strains using DNA templates from Mycobacterium tuberculosis H37Rv (MT) strain number 27294 from American Type Culture Collection (ATCC) USA, and Mycobacterium avium subspecies paratuberculosis (MAP) strain number 103963 from Collection de l’ Institute Pasteur (CIP) France.
Lane 1: 100 bp DNA ladder (Fermentas, Catalogue No. SM1153); Lanes 2 and 3: DNA from MT ATCC strain and MAP CIP strains amplified with primers sets corresponding to 16S rRNA 1429; Lanes 4 and 5: DNA from MT and MAP strains amplified with primer sets corresponding to katG 463; lanes 6 and 7 : DNA from MT and MAP strains amplified with primer sets corresponding to hsp65 631; lanes 8 and 9: DNA from MT and MAP strains amplified with primer sets corresponding to gyrA 95; lanes 10 and 11: negative control - Nuclease free water (Merck, Catalogue No. 612151181001730)
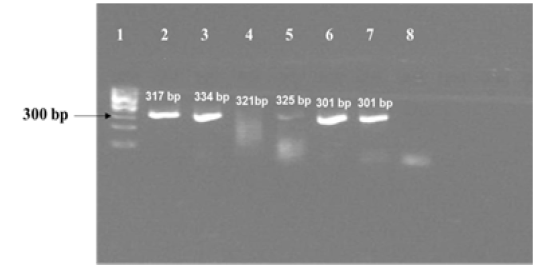
Figure 1c: Results of PCR using primers for sequencing experiments on amplicon templates greater than 300 bp. Corresponding forward and reverse primer sets for 16S rRNA 1429, katG 463 , hsp 65 631, gyrA 95, gyrB (1410) and gyrB (675) gene targets were used for amplification of PCR products of expected sizes of 317 bp, 334 bp, 321 bp, 325 bp, 301 bp and 301 bp respectively.
Lane 1: 100 bp DNA ladder (Fermentas, Catalogue No. SM1153); lanes 2 to 7: DNA template from MT ATCC strain was amplified using primer sets corresponding to nucleotide sequence of 16S rRNA 1429, katG 463, hsp 65 631, gyrA 95, gyrB (1410) and gyrB (675); Lane 8: Negative control–Nuclease free water (Merck, Catalogue No. 612151181001730
The nine field isolates of Mycobacterium were amplified by PCR with the gene targets gyrA 95 and gyrB (1410) (Fig 2a & 2b). In addition, partial sequences of the above isolates were obtained from amplification of gyrA and gyrB gene sequences that were greater than 300 bp using sequencing primers (Table 2). These partial sequences were matched with Mycobacterium sequence available in the public domain by BLASTn analysis. Sequence data of 8/9 isolates matched with M. tuberculosis H37Rv reference strain, while 1/9 isolate showed homology with sequence of M. intracellulare. A total of 17 partial sequences were generated from gyrA and gyrB were submitted to GenBank; the GenBank Accession number has been provided in Table 2. Sequence homologies among field strains and reference strains were supported by disparity index test results (Table 4a and 4b), and the phylogenetic tree constructed based on partial sequences of gyrB, also clustered 8 isolates with MT reference strain and 1 isolate with M.I (Figure 3).
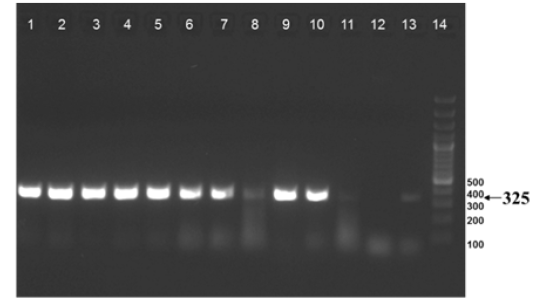
Figure 2a: Amplification of gyrA 95 gene from DNA extracted from cultures of field samples and reference strains.
Lanes 1 -8: Field isolates of Mycobacterium tuberculosis – Isolate No. IS1 to IS8; Lanes 9 -10: Mycobacterium tuberculosis H37Rv - American Type Culture Collection (ATCC) reference strain 27294; Mycobacterium bovis BCG ATCC 35734; Lanes 11-12: Water as negative control; Lane 13: Field isolate No. IS 9 of Mycobacterium. Lane 14: Ladder 100bp (Fermentas; Catalogue number SM1153)
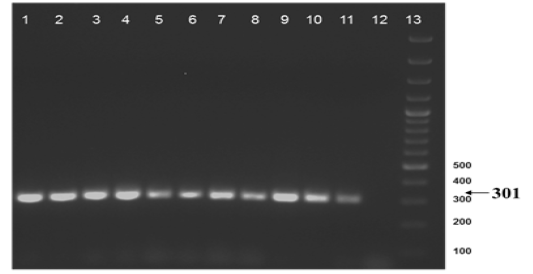
Figure 2b: Amplification of gyrB (1410) gene from DNA extracted from cultures of field samples and refernce strains.
Lane 1 to 8: Field isolates of Mycobacterium tuberculosis - Isolate No. ISI to IS8; Lane 9: Field isolate No. IS 9 of Mycobacterium; Lane 10-11: M. tuberculosis H37Rv ATCC 27294; M. bovis BCG ATCC 35734; Lane 12: Water as negative control; Lane 13: Ladder 100bp (Fermentas; Catalogue number SM1153)
Primers and probes were designed to develop Real-Time PCR and to amplify the gyrA and gyrB genes harboring strain specific SNP regions for discrimination of M. tuberculosisand M. bovis. The analytical specificity of Real-Time PCR with gene targets gyrA and gyrB was evaluated by amplifying Mycobacterium tuberculosis H37Rv (ATCC 27294), Mycobacterium bovis BCG Pasteur (ATCC 35734), Mycobacterium avium subspecies paratuberculosis (CIP 103963), Mycobacterium avium subspecies paratuberculosis (ATCC 19698), Campylobacter fetus subspecies fetus (ATCC 27374), Campylobacter fetus subspecies venerealis (ATCC 19438), Brucella abortus 544 (ATCC 23448), Brucella abortus S19
Table 4a: Gyrase A Disparity Index Test
|
Mycobacterium tuberculosis H37Rv |
Mycobacterium tuberculosis ATCC 27294 |
IS-1 | IS-2 | IS-3 | IS-4 | IS-5 | IS-6 | IS-7 | IS-8 | |
|
Mycobacterium tuberculosis H37Rv |
- | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
|
Mycobacterium tuberculosis ATCC 27294 |
1.000 | - | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| IS-1 | 1.000 | 1.000 | - | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| IS-2 | 1.000 | 1.000 | 1.000 | - | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| IS-3 | 1.000 | 1.000 | 1.000 | 1.000 | - | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| IS-4 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | - | 0.000 | 0.000 | 0.000 | 0.000 |
| IS-5 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | - | 0.000 | 0.000 | 0.000 |
| IS-6 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | - | 0.000 | 0.000 |
| IS-7 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | - | 0.000 |
| IS-8 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
- |
ATCC: American Type Culture Collection
Table 4b: Gyrase B Disparity Index Test
|
Mycobacterium tuberculosis H37Rv* |
Mycobacterium tuberculosis ATCC 27294 |
Mycobacterium intracellulare ATCC 13950 |
IS-1 | IS-2 | IS-3 | IS-4 | IS-5 | IS-6 | IS-7 | IS-8 | IS-9 | |
|
Mycobacterium tuberculosis H37Rv |
- | 0.000 | 0.408 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.056 |
|
Mycobacterium tuberculosis ATCC 27294 |
1.000 | - | 0.408 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.056 |
|
Mycobacterium intercellulare ATCC 13950 |
0.008 | 0.006 | - | 0.408 | 0.408 | 0.408 | 0.408 | 0.408 | 0.408 | 0.408 | 0.408 | 0.060 |
| IS-1 | 1.000 | 1.000 | 0.006 | - | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.056 |
| IS-2 | 1.000 | 1.000 | 0.006 | 1.000 | - | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.056 |
| IS-3 | 1.000 | 1.000 | 0.006 | 1.000 | 1.000 | - | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.056 |
| IS-4 | 1.000 | 1.000 | 0.004 | 1.000 | 1.000 | 1.000 | - | 0.000 | 0.000 | 0.000 | 0.000 | 0.056 |
| IS-5 | 1.000 | 1.000 | 0.012 | 1.000 | 1.000 | 1.000 | 1.000 | - | 0.000 | 0.000 | 0.000 |
0.056 |
| IS-6 | 1.000 | 1.000 | 0.008 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | - | 0.000 | 0.000 | 0.056 |
| IS-7 | 1.000 | 1.000 | 0.006 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | - | 0.000 | 0.056 |
| IS-8 | 1.000 | 1.000 | 0.006 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | - | 0.056 |
| IS-9 | 0.244 | 0.226 | 0.070 | 0.232 | 0.218 | 0.230 | 0.238 | 0.256 | 0.218 | 0.230 | 0.188 |
- |
ATCC: American Type Culture Collection
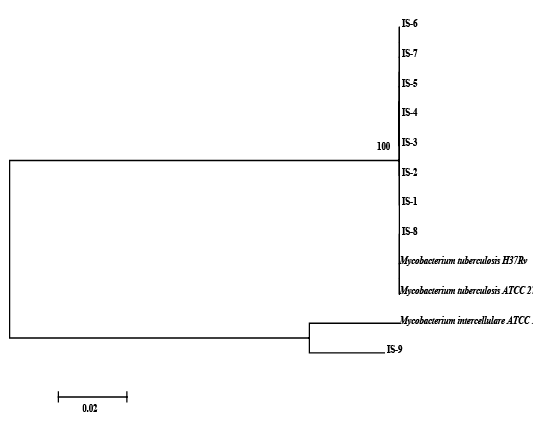
Figure 3: Relationship among field strains of Mycobacterium of animal origin with respect to reference strains compared on the basis of homology of sequences in the region of gyrB represented in the form of a Phylogenetic tree
(USDA/ Mar. 98), Saccharomyces cerevisiae (Thermofisher Scientific Cat. No. C81000), Mycobacterium phlei (ATCC 106789), Mycobacterium smegmatis (ATCC 607), Tritrichomonas fetus (ATCC 30003) as described in Table 3. The result obtained showed that gyrA and gyrB gene targets are specific since it detected only M. tuberculosis and M. bovis standard reference strains, respectively (Table 3). Further, for both gene targets [gyrA 95 and gyrB (1410)] the Real-Time PCR results did not differ significantly from culture as determined Student’s t-test (Table 5a and 5b).
The Real-Time PCR of gyrA 95 could detect up to 450 fg of M. tuberculosis DNA from templates generated by tenfold serial dilutions of DNA, ranging from 45ng to 45 agrun in triplicates. The Cq value was determined to be 41.57 by running negative samples up to 50 cycles. Likewise the real time PCR of gene target gyrB (1410) was able to detect up to 600 fg of M. bovis DNA from the templates generated by ten-fold serial dilutions of DNA, ranging from 60 ng to 60 ag run in triplicates. To declare sample as positive the Cq value was determined as 41.22 by running a negative sample up to 60 cycles.
Further, the ROC analysis revealed the ASe of the gyrA 95 Real-Time PCR was 100% at 95% Confidence Intervals (CI) |66.40 – 100.00|; and Asp 100% at 95% CI |75.30 – 100.0|, respectively (Table 6). The repeatability of gyrA 95 and gyrB (1410) Real-Time PCR determined by comparing data of replicates [replicate: 1 to 2; 1 to 3; and 2 to 3] by Bland Altman Plot, indicated that the variation of
Table 5a: Analysis of Specificity of gyrA 95 Real time PCR by Student’s t-test
| Independent Samples t-test | ||
| Parameters | Variables | |
| Real time PCR | Culture | |
|
Sample size |
22 |
22 |
|
Arithmetic mean |
0.4091 |
0.4091 |
|
95% CI for the mean |
0.1860 to 0.6322 |
0.1860 to 0.6322 |
|
Variance |
0.2532 |
0.2532 |
|
Standard deviation |
0.5032 |
0.5032 |
|
Standard error of the mean |
0.1073 |
0.1073 |
|
F-test for equal variances |
P = 1.000 |
|
| T-test (assuming equal variances) | ||
| Parameters | Values | |
|
Difference |
0.0000 |
|
|
Pooled Standard Deviation |
0.5032 |
|
|
Standard Error |
0.1517 |
|
|
95% CI of difference |
-0.3062 to 0.3062 |
|
|
Test statistic t |
0.000 |
|
|
Degrees of Freedom (DF) |
42 |
|
|
Two-tailed probability |
P = 1.0000 |
|
| Residuals | ||
|
D'Agostino-Pearson test for Normal distribution
|
Reject Normality (P<0.0001) | |
Table 5b: Analysis of Specificity of gyrB (1410) Real time PCR by Student’s t test
| Independent Samples t-test | ||
| Parameters | Variables | |
| Real time PCR | Culture | |
|
Sample size |
22 |
22 |
|
Arithmetic mean |
0.04545 |
0.04545 |
|
95% CI for the mean |
-0.04907 to 0.1400 |
-0.04907 to 0.1400 |
|
Variance |
0.04545 |
0.04545 |
|
Standard deviation |
0.2132 |
0.2132 |
|
Standard error of the mean |
0.04545 |
0.04545 |
|
F-test for equal variances |
P = 1.000 |
|
| T-test (assuming equal variances) | ||
| Parameters | Values | |
|
Difference |
0.0000 |
|
|
Pooled Standard Deviation |
0.2132 |
|
|
Standard Error |
0.06428 |
|
|
95% CI of difference |
-0.1297 to 0.1297 |
|
|
Test statistic t |
0.000 |
|
|
Degrees of Freedom (DF) |
42 |
|
|
Two-tailed probability |
P = 1.0000 |
|
| Residuals | ||
|
D'Agostino-Pearson test for Normal distribution |
reject Normality (P<0.0001) |
|
Table 6: Determination of Analytical Specificity (ASp) and Analytical Sensitivity (ASe) of gyrA 95 Real time PCR by ROC Analysis
| Area under the ROC curve (AUC) | ||||||
| Parameters | Values | |||||
|
Area under the ROC curve (AUC) |
1.000 |
|||||
|
Standard Errora |
0.000 |
|||||
|
95% Confidence intervalb |
0.846 to 1.000 |
|||||
|
Significance level P (Area=0.5) |
<0.0001 |
|||||
| Youden index | ||||||
| Parameters | Values | |||||
|
Youden index J |
1.0000 |
|||||
|
Associated criterion |
>0 |
|||||
|
Sensitivity |
100.00 |
|||||
|
Specificity |
100.00 |
|||||
| Criterion values and coordinates of the ROC curve | ||||||
|
Criterion |
Sensitivity |
95% CI |
Specificity |
95% CI |
+LR |
-LR |
|
≥0 |
100.00 |
66.4 - 100.0 |
0.00 |
0.0 - 24.7 |
1.00 |
|
|
>0 |
100.00 |
66.4 - 100.0 |
100.00 |
75.3 - 100.0 |
0.00 |
|
|
- |
0.00 |
0.0 - 33.6 |
100.00 |
75.3 - 100.0 |
1.00 |
|
Figure depicted in bold under the column ‘Criterion’ and shown in the row mentioning the condition ‘>0’ was considered for sensitivity and specificity of the assay at 95% confidence intervals.
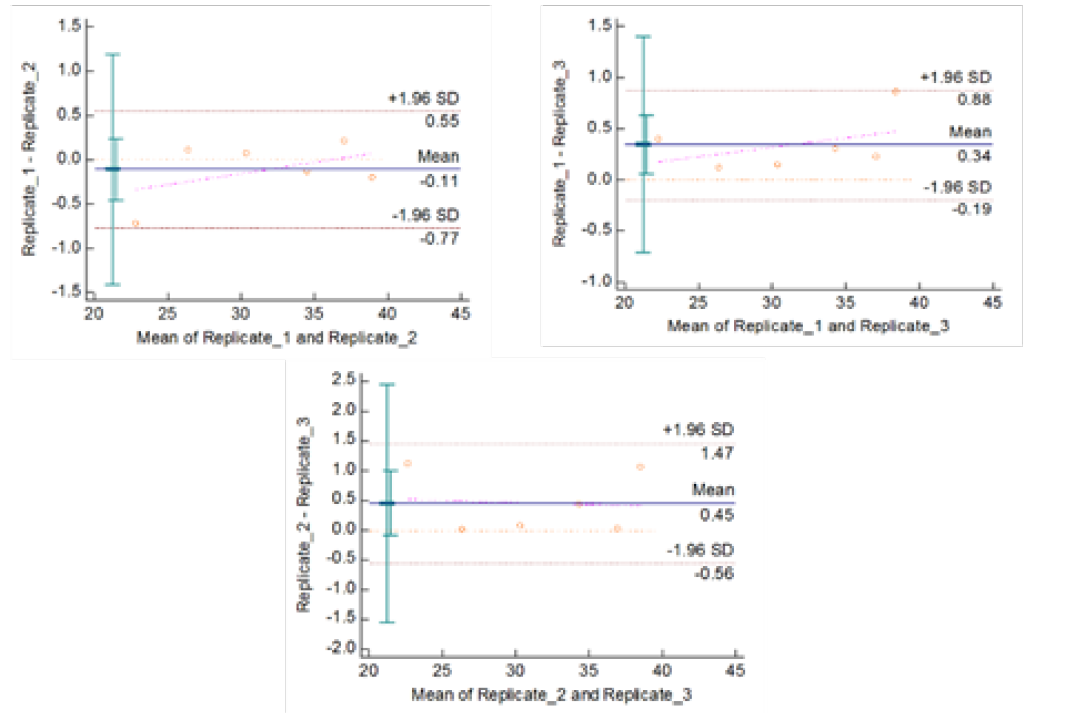
Figute 4b: Repeatability of gyrA 95 Real Time PCR: Variation in mean Cq values between replicates analysed by Bland-Altman Plot
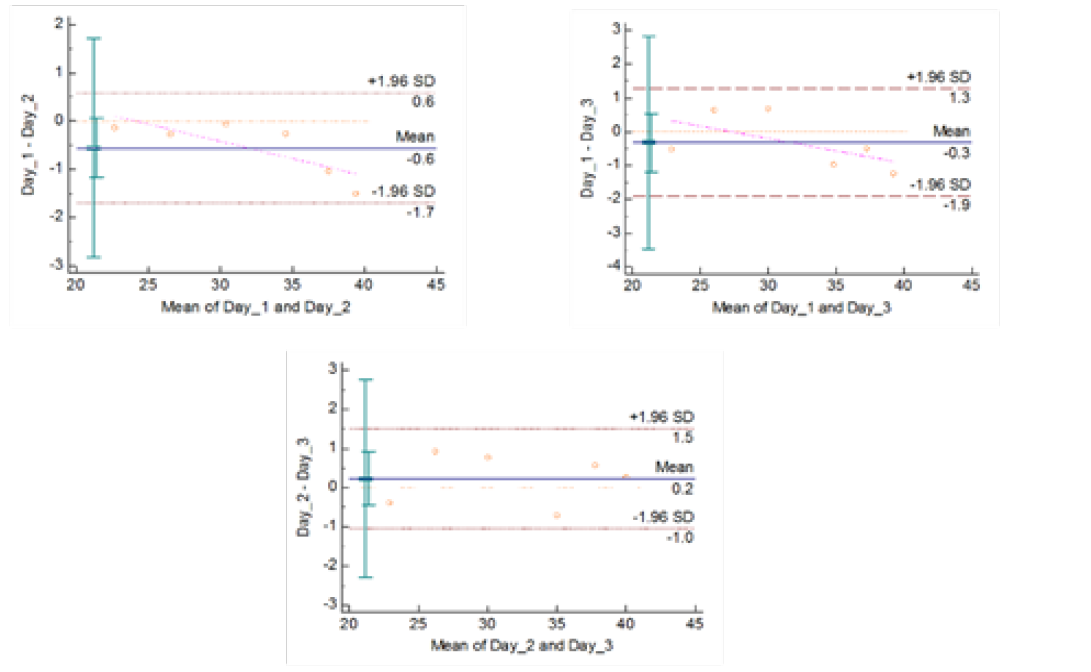
Figure 4c: Reproducibility of gyrA 95 Real Time PCR: Variation in mean Cq values between days analysed by Bland-Altman Plot
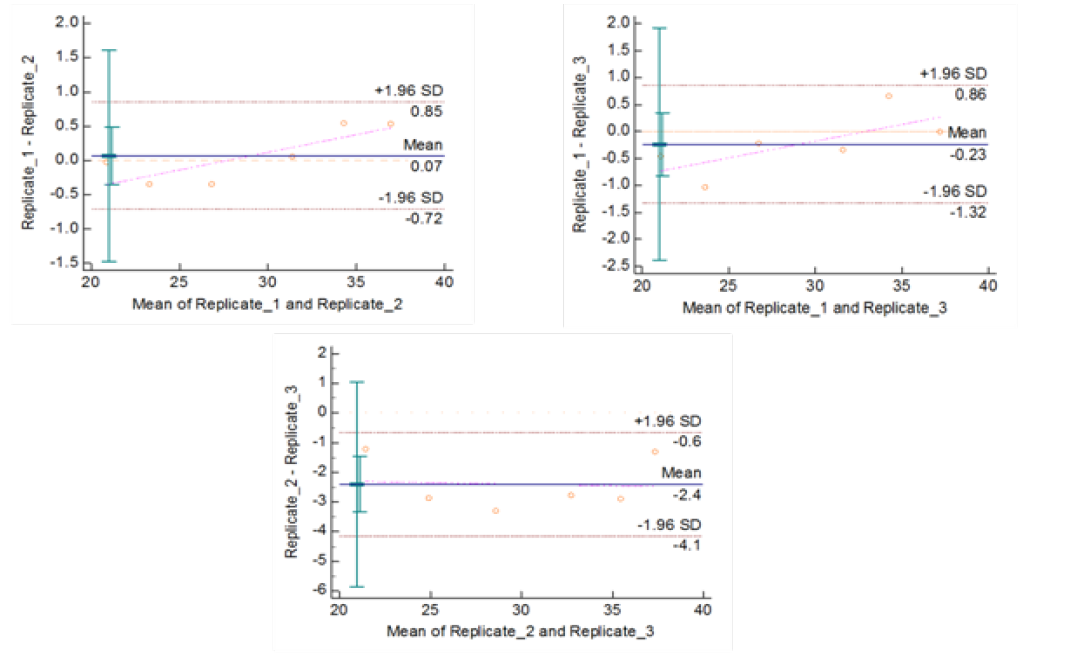
Figure 5b: Repeatability of gyrB (1410) Real Time PCR: Variation in mean Cq values between replicates analysed by Bland-Altman Plot
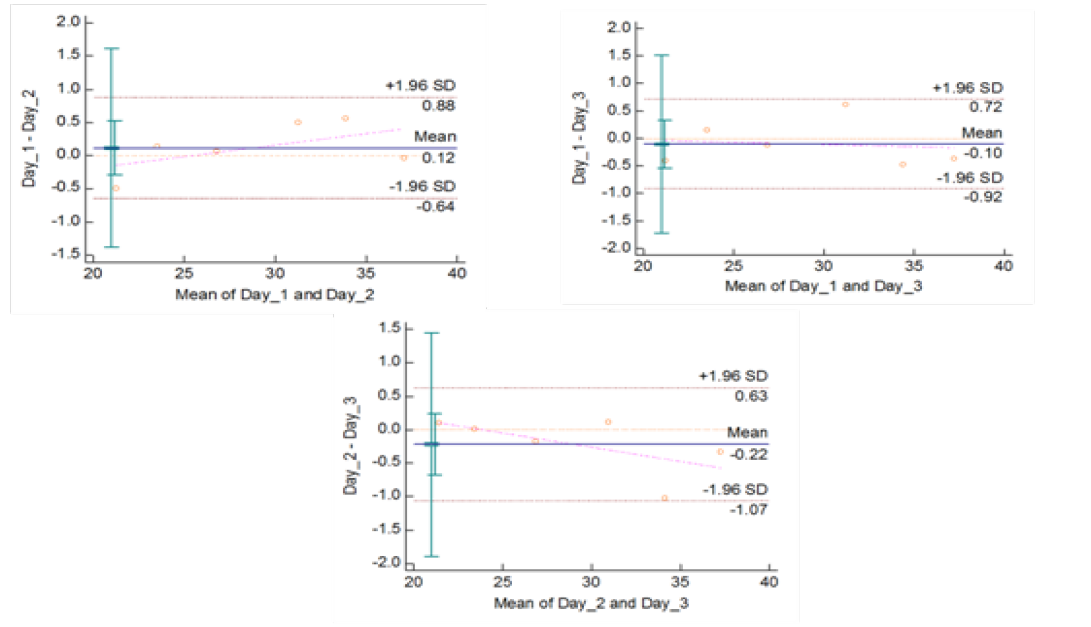
Figure 5c: Reproducibility of gyrB (1410) Real Time PCR: Variation in mean Cq values between days analysed by Bland-Altman Plot
means of replicates lay within acceptable statistical range of + 1.96 of Standard Deviation (SD) (Fig 4a and 4b). The reproducibility of gyrA 95 and gyrB (1410) Real-Time PCR determined by comparing the day-wise data of replicates [Day: 1 with 2; Day 1 with 3; and Day 2 with 3] by Bland Altman Plot, indicated that the variation of means of replicates lay within acceptable statistical range of + 1.96 of Standard Deviation (SD) (Fig 5a and 5b).
All the isolates of mycobacterium were confirmed by single-tube-nested PCR when a positive amplification was of 220 bp followed by a nested amplification of a 123 bp amplicon from the IS6110 region was obtained. Further analysis/ characterisation using by GenoType® Mycobacterium CM (Common Mycobacteria) and GenoType® MTBC (Mycobacterium tuberculosis complex), deduced as M. tuberculosis (8) on the basis of hybridization pattern generated, and one as M. intracellulare.
DISCUSSIONS
Although bovine tuberculosis in members of the Bovidae family is generally due to infection with MB (OIE, 2014), they are also infected with species classified as Mycobacterium other than tuberculosis (MOTT) (Romero et al., 2008; Gortazar et al., 2011), and harbor co-infection with M. avium (Thorel et al., 2001; Gortazar et al., 2011) and with M. tuberculosis (Mishra et al., 2005; Jia et al., 2012). This necessitates accurate detection of the pathogen. Identification of pathogens by culture, intra-dermal skin test and IFN-γ assay have various limitations (OIE, 2009; Faye et al., 2011; Alvarez et al., 2012; Bassessar et al., 2014). Hence the development and use of molecular diagnostic tools for rapid and specific detection of infection assumes significance. Molecular techniques such as the use of rapid, specific and sensitive diagnostic Real Time PCR are being increasingly applied under clinical settings for arriving at a quicker and more accurate evaluation of the status of tuberculosis infection. Molecular identification of strains belonging to the genus Mycobacterium classified under MTBC group for differentiation of Mycobacterium tuberculosis strains of species and lineage level, and to study their evolution, based on variation in the genome due to SNPs as been reported earlier (Sreevatsan et al., 1997; Brosch et al., 2002; Filliol., 2006; Bouakaze et al., 2010; Bouakaze et al., 2011). This approach has been extended to monitor presence of virulent MT strains such those belonging to the Beijing lineage (Alonso et al., 2010) and rapidly identify drug resistant strains (Ramirez et al., 2010).
Realizing the potential of this approach based on the above literature, we had set about investigating whether certain gene targets such as those containing the SNPs (Hsp65 631, katG 463, 16S rRNA 1429, gyrA 95, gyrB (675), gyrB (756) and gyrB (1410) reported earlier, that have been found suitable and specific for differentiation of strains within the MTBC, were suitable for development of discriminatory Real Time PCRs. The above reports had described the SNaPshot mini-sequencing approach for strain differentiation. In this work we wanted to study whether differences in these genes due to SNPs could be adopted for development of Real Time PCRs that exploits minor groove binding probes containing these SNPs, flanked by forward and reverse primers encompassing the selected regions within a specific genome.
The initial assays reported here were designed to test whether all the eight regions harboring the SNPs could be specifically amplified in a simple gel PCR format and prove the actual existence of these specific SNPs within sets of amplicons (one set containing amplicons less than 300 bp, and the other set with amplicons greater than 300 bp in size) by using sequencing data from these amplicons. The sequencing information was then used to determine the extent of homology of these sequences with published sequence information for the M. tuberculosis H37Rv and M. bovis available in the NCBI database. Using the above exercise for quality assurance, we further designed differentially labeled fluorescent minor groove binding probes and corresponding primer pairs for development of discriminatory real time PCRs. We limited the assessment of our designs, by checking the suitability of our assays concentrating only on two SNP containing gene targets–gyrA 95 and gyrB (1410).
Although we were able to generate specific amplicons of sizes greater than 300 bp [317 bp from 16S rRNA 1429, 334 bp from katG 463, 321 bp from hsp65 631, 325 bp from gyrA 95, 301 bp from gyrB (1410) and 301 bp from gyrB (675) respectively, as shown in Fig 1c] from six out eight selected gene targets using sequencing primers designed for this study (Table 1), we were not able to sequence and use them further, for the development Real Time PCR assay formats, because of time constraints. As sequencing of purified PCR products (< 300 bp) derived from amplification of eight gene targets from DNA of reference strain of MT had indicated the presence of SNPs specific to these gene targets, it is suggested that the corresponding MGB probes designed in this study probably have the potential for development of separate Real Time PCR assays, that can discriminate MT from M. canetti, M. pinnipedii, M. microti and M. caprae in future.
Assessment of our results employing DNA templates from reference MTBC strains and nuclease free water as positive and negative controls respectively, non-Tuberculous Mycobacterium strains and other non-Mycobacterium strains, indicated that the assays containing the VIC labeled probe employing the SNP in gyrA 95 was specific for detection of M. tuberculosis strain, while the FAM labeled SNP gyrB (1410) probe was specific for detection of M. bovis strains; and both assays could be completed in 78 minutes, and were repeatable and reproducible. As expected the FAM labeled SNP gyrA 95 probenon-specifically detected all the members belonging to the MTBC complex. Similarly, the VIC labeled SNP gyrB (1410) probe detected all the members belonging to the MTBC complex. The results of PCR assays validated the specificity of selection of primers and differentially labeled fluorescent MGB probes used in this study; thus confirming the authenticity of our initial in silico sequence analysis.
Further validation of our discriminatory SNP Real Time PCR assays was established when strains previously identified by culture and commercial hybridization methods showed complete concordance in this study; analysis of partial sequencing data using bioinformatics tools generated from amplification of gyrA 95 and gyrB (1410) from 9 Indian field isolates of animal origin correctly delineated eight strains as MT and one as MI, but none as MB. These results taken together were encouraging since they exhibited the potential for further validation of discriminatory SNP based diagnostic Real Time PCRs to be explored under clinical settings. As an extension of this study, future program could be aimed at determining the diagnostic estimates for these assays using DNA templates directly from clinical samples; since in a recent study (Wampande et al., 2015) it has been emphasized that SNP Real Time PCR possess the potential to rapidly and specifically detect 70 MTBC strains to lineage and sub-lineage specific accuracy, employing a throughput format.
Although the present study indicated that SNP Real Time PCR was sensitive enough to detect 450 fg of MT and 600 fg of MB, the assay can be upgraded by inclusion of an internal amplification control (IAC) in tests for determination of the copy number and therefore upgrade it into a quantitative assay format. The IAC can serve as additional quality assurance indicators (Mukherjee et al., 2015). The efficiency and robustness of these assays need to be also tested in future by evaluating performances using DNA templates generated from diverse clinical matrices. The development of assays reported here is restricted to discrimination of strains within the MTBC. However, they are unable to distinguish lineages or sub-lineages within the MTBC, since our recent spoligo typing analysis of 8 MT field isolates from India revealed the prevalence of 5 distinct lineages in the sub-continent [Mukherjee et al., – manuscript accepted for publication in Revue Scientifique et Technique Vol. 37 (3)], this limitation seems to restrict the repertoire of the above assays.
Comparison of sequencing data from amplicons of sizes 72 bp, 72 bp, 120 bp, 198bp, 123 bp, 271 bp, 167 bp and 150 bp generated from amplification of gene target of hsp65 631, katG 463, 16S rRNA1429, gyrA 95, gyrB (675), gyrB(756), gyrB (1410) and gyrB (675+756) respectively, from DNA template of MT H37Rv reference strain; and amplicons of size 325 bp and 301bp generated from the gyrA 95 and gyrB (1410) gene targets from DNA templates of eight Mycobacterium field isolates, indicated that the alleles in the SNPs for each selected targets were same as those indicated in the published genome sequence information (GenBank Accession No. NC_000962.3). In this analysis, the presence of the allele ‘G’ in the sequence (CGG) of katG 463 and ‘G’ in the sequence (AGC) of gyrA 95 in the reference and field strains seems to suggest that field strains of MT belong to PGG 3, and further the presence of the allele ‘C’ in gyrB (1410) and absence ‘T’ confirmed the field strains were not of M. bovis or M. bovis BCG. However, the gyrA 95 VIC labeled probe would not be suitable for identification MT strains belonging to PPG 1b because the allele in such strains is ‘C’ which is also present in other members of the MTBC. Earlier reports (Brosch et al., 2002; Bouakaze et al., 2011) based on testing of a total of 81 field MT strains had indicated that 20% to 35% belonged PGG 1b, and 65 to 80% of these strains belonged to PGG 2 and 3; of which 4% were identified as PGG 3. So it seems that MT field strains assigned to PGG 3 are rare and their recovery from animals in India might be pointing towards an important link related to the acquirement of MT infection by animals from the primary human host for this pathogen by a ‘spill over’ mechanism. ‘Spill over’ mechanism as a method of spread of MT infection from human to animals have been suggested earlier (Malama et al., 2014).
Although, the analysis of partial sequencing data of MT and MI strains by Disparity Index Test for gyrA and gyrB and the profile of Phylogenetic Tree based on gyrB, indicated complete homology of field strains with reference strains, and a synonymous substitution of SNP in gyrA 95 and gyrB (1410); we were not sure whether these mutations were naturally acquired, since our observations for the presence SNPs on field strains were limited to study of only a part of the two genomes - gyrA and gyrB and not for entire genome (Kasai et al., 2000; Ilina et al., 2013; Oudghiri et al., 2018). Also, in order to conclude that mutations in SNPs have been naturally acquired, it has been recommended that a larger number of housekeeping genes be considered for investigation (Ilina et al., 2013; Oudghiri et al., 2018), since we focused our investigation for field strains on two gene targets, it is difficult to make a final conclusion about the evolution of mutation in these field strains.
ACKNOWLEDGEMENTS
The authors are grateful to the management of the National Dairy Development Board (NDDB), Anand, for providing the facility to carry out this work.
AUTHORS CONTRIBUTION
VSB carried out all the optimization and validation work and prepared the data. FM conceptualized and designed the study, did in silico analysis of sequence and wrote the paper. BM helped in the designing of probes. SYP performed the initial PCR on seven gene targets. AP and SKR collected the original field samples. KSNLS analyzed the amplicon sequence information of gyrA 95 and gyrB (1410) SNP Real Time PCRs of field isolates using disparity index test and creation of phylogenetic tree. GKS provided the administrative and financial support from the NDDB. All the authors are thankful to VAS for critically reviewing and editing the manuscript. Special thanks are due for Caroline Bouakaze for critically examining the authenticity of the probes designed in silico by FM.
CONFLICT OF INTEREST
The authors declare no conflict of interest related to this article.
REFERENCES