Journal of Infection and Molecular Biology
Research Article
Molecular Characterization and Phylogenetic Analysis of Seven Flavobacterium columnare Strains Isolated from Freshwater Fishes of Eastern India
Soumya Sankar Rath1*, Pragyan Roy2, Santi Lata Sahoo1, Basanta Kumar Das2
1Department of Applied Microbiology, Utkal Univeristy, Bhubaneswar-751004, India; 2Fish Health Management Division, Central Institute of Freshwater Aquaculture, Kausalyaganga, Bhubaneswar-751002, India.
Abstract | Flavobacterium columnare is the causative agent of columnaris disease in most of the freshwater fishes. In the present study, molecular and phylogenetic analysis were used to demonstrate the genetic diversity among F. columnare strains isolated from freshwater fishes e.g. Catla catla, Labeo rohita, Anabas testudineus, Carassius auratus. A prominent band of 1.5 Kbp was observed for all the seven strains of F. columnare. A phylogenetic tree was constructed with the 16S rDNA sequences of these seven strains with other published F. columnare strain sequences available in the GenBank. Sequence identity and phylogenetic study revealed F. columnare strains isolated form Catla catla were closely related to each other and strains isolated from Labeo rohita, Anabas testudineus, Carassius auratus were distantly related to each other. In conclusion, F. Columnare strains infecting freshwater fishes were different to each other as revealed by 16S rDNA and phylogenetic studies.
Keywords | Flavobacterium columnare, Catla, Rohu, 16S rDNA gene, Phylogenetic tree
Editor | Tahir Yaqub, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | October 25, 2016; Accepted | November 01, 2016; Published | November 07, 2016
*Correspondence | Soumya Sankar Rath, Department of Applied Microbiology, Utkal University, Bhubaneswar-751004, Odisha, India; Email: soumya.rath21@gmail.com
Citation | Rath SS, Roy P, Sahoo SL, Das BK (2016). Molecular characterization and phylogenetic analysis of seven Flavobacterium columnare strains isolated from freshwater fishes of eastern India. J. Inf. Mol. Biol. 4(2): 16-23.
DOI | http://dx.doi.org/10.14737/journal.jimb/2016/4.2.16.23
ISSN (Online) | 2307-5465; ISSN (Print) | 2307-5716
Copyright © 2016 Rath et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Fishes are continuously exposed to a wide range of microorganisms present in the environment and columnaris disease is one of the most widespread bacterial diseases in aquaculture industry. Flavobacterium columnare is a frequently occurring bacterial pathogen in freshwater fishes and is the causative agent of columnare disease (Plumb, 1994). It is a Gram negative rod (2-10 µm) bacterium and affects most of the freshwater fishes in eastern India (Das et al., 2009).
The PCR based techniques employing universal primers have been widely used for the phylogenetic analysis and taxonomic identifications (Fox et al., 1977; Tringe et al., 2005). The 16S rDNA gene sequencing is one of the most effective techniques to differentiate F. columnare from other species of Flavobacteria (Darwish and Ismaiel, 2005; Toyama et al., 1996). The 16S rDNA gene is a highly conserved gene in prokaryotic organisms that has been used in phylogenetic analysis to provide taxonomic information (Weisburg et al., 1991; Triyanto et al., 1999). The first six hundred nucleotides in the 5` terminus of the 16S rDNA contains enough information to allow accurate alignment of bacterial sequences to the main lines of descent and this terminus has been recommended as the region of interest for molecular analysis (Lieasck et al., 1997). This helps for the effective disease diagnosis and treatment of the causal agent by identifying the organism from its species level. Morphological and biochemical tests are the primary methods of diagnosis of fish diseases (Elkamel and Mohamed, 2012). However, these methods often lack specificity and many fish pathogens are difficult to detect. For this reason the 16S rDNA sequencing technique is used for the rapid and confirmatory identification of the organism as well as the phylogenetic analysis among different strains of same species.
In the present study, the fish pathogen, F. columnare were isolated from different infected fishes i.e. Indian major carp rohu, Labeo rohita; catla, Catla catla; anabas, Anabas testudineus and goldfish, Carassius auratus and were identified based on the 16S rDNA sequencing and accordingly phylogenetic analysis were done. The genetic diversity of these F. columnare strains were investigated analyzing the phylogenetic tree also the relationship among the more closely related strains was tested.
Materials and Methods
Bacterial Strain
Seven F. columnare strains used in this study were isolated from different fish species of eastern India (Table 1). All the seven strains were maintained in the Fish Health Management Division, Central Institute of Freshwater Aquaculture (CIFA, India) and possess all the biochemical characteristics described by Griffin (1992).
Table 1: F. columnare strains from different organs of catla, rohu, goldfish and anabas with accession number
|
Bacterial Strain No. |
Host |
Organ of Isolation |
Accession Number |
|
CFCCO41 |
Catla |
Operculum |
KF051085 |
|
CFCRVB43 |
Rohu |
Ventral belly |
KF051086 |
|
CFCGFG50 |
Gold fish |
Gill |
KF051087 |
|
CFCRG55 |
Rohu |
Gill |
KF051088 |
|
CFCCG62 |
Catla |
Gill |
KF051089 |
|
CFCCSL66 |
Catla |
Skin lesions |
KF051090 |
|
CFCACR72 |
Anabas |
Caudal region |
KF051091 |
Genomic DNA Isolation
The genomic DNA was extracted using MB505 HiPurATM Bacterial and yeast genomic DNA purification kit (Himedia Ltd., Mumbai, India) following the instruction provided by the manufacturer. Bacterial culture (1.5 ml) was taken from 24 h broth culture in an eppendrof tube and centrifuged (Jouan-DR4, USA) at 10,000 rpm for 5 min. The pellet collected were resuspended in180 μl of Lysis solution AL and 20 μl of RNase A followed by incubation of 2 min at room temperature. Proteinase K (20 mg/ml) of 20 μl was added to the solution and incubated for 30 min at 55°C. Then 200 μl of Lysis solution C1 was added, mixed properly and incubated at 55°C for 10 min, followed by addition of 200 μl 95-100% ethanol. The entire contents of the tube were inserted onto binding column and centrifuged at 10,000 rpm for 1 min. The column was placed in a fresh 2 ml collection tube, then 500 μl of prewash solution-PWB was added and centrifuged at 10,000 rpm for 1 min. After that 500 μl of wash solution was added to the column and centrifuged at 10,000 rpm for 1 min. Then the column was placed in another new 2 ml collection tube followed by addition of 200 μl of Elution buffer-ET and incubated at room temperature for 2 min. The column was then centrifuged at 10,000 rpm for 1 min to elute DNA. The purity of the DNA was checked by spectrophotometer (BIORAD-Smart Spec. 3000, Germany).
PCR Amplification of 16S rDNA Gene
The 16S rDNA gene was amplified by using 16S rDNA forward primer and 16S rDNA reverse primer. The universal primer, forward primer (5’-AAGAGTTTGATCCTGGCTCAG-3’) and reverse primer- (5’-GGTTACCTTGTTACGACTT- 3’) obtained from Bangalore Genei (Bangalore, India) were used to amplify the 16S rDNA sequences. The PCR was performed using an automatic programmable thermal cycler (Applied Biosystem, USA). The PCR reaction mixture (25µl) was prepared which includes 17.25 µl nuclease free water, 2.5 µl assay buffer (Bangalore Genei, Bangalore, India), 0.25 µl Magnesium chloride (Bangalore Genei, Bangalore, India), 1 µl dNTPs (Bangalore Genei, Bangalore, India), 1 µl 16S forward primer and 1 µl, 16S reverse primer, 0.8µl Taq DNA polymerase (Bangalore Genei, Bangalore, India) and 1.2 µl genomic DNA. The PCR condition included a preheating of 95°C followed by 35 cycles of denaturation at 94°C for 1 minute, annealing at 58°C for 1 minute, extension at 72°C for 1 minute and final delay of 72°C for 10 minutes. The PCR amplified products were subjected to electrophoresis in 1.2% agarose gel containing 2.5µl ethidium bromide with an expected size of 1.5 kb, using standard molecular weight marker of 1 kbp DNA ladder (Bangalore Genei, India). The gel was then photographed by using Gel documentation (Gel Doc- ITTM Imaging System, Germany). PCR amplicons were purified by using HiPuraTM PCR purification kit and sequenced at EUROFIN laboratories Pvt. Ltd. Bangalore, India.
Sequence Analysis
The 16S rDNA gene sequences obtained from the sequencing were analyzed with the other F. columnare strain 16S rDNA gene sequence available in GenBank with the BLAST (Basic Local Alignment Sequencing Tool) algorithm provided by NCBI (National Centre for Biotechnology Information).
Phylogenetic Analysis
The seven F. columnare 16S rDNA sequences were aligned using the online multiple alignment software CLUSTALW. MEGA 5.10 software was used to construct the phylogenetic tree. The phylogenetic tree was constructed by using Neighbor-Joining method (Saitou and Nei, 1987) with other published sequences available at GenBank. The evolutionary distances were computed using the Maximum Composite Likelihood Method (Tamura et al., 2004).
Table 2: Score of pair wise alignment of sequences obtained from 16S rDNA of F. columnare strains isolated from eastern India
|
Sequence A |
Name |
Length |
Sequence B |
Name |
Length |
Score |
|
1 |
KF051086 |
1155 |
2 |
KF051087 |
1157 |
78.87 |
|
1 |
KF051086 |
1155 |
3 |
KF051089 |
1151 |
81.06 |
|
1 |
KF051086 |
1155 |
4 |
KF051090 |
1164 |
82.08 |
|
1 |
KF051086 |
1155 |
5 |
KF051091 |
1167 |
81.82 |
|
1 |
KF051086 |
1155 |
6 |
KF051088 |
1216 |
81.47 |
|
1 |
KF051086 |
1155 |
7 |
KF051085 |
1128 |
81.91 |
|
2 |
KF051087 |
1157 |
3 |
KF051089 |
1151 |
92.79 |
|
2 |
KF051087 |
1157 |
4 |
KF051090 |
1164 |
94.3 |
|
2 |
KF051087 |
1157 |
5 |
KF051091 |
1167 |
93.09 |
|
2 |
KF051087 |
1157 |
6 |
KF051088 |
1216 |
93.26 |
|
2 |
KF051087 |
1157 |
7 |
KF051085 |
1128 |
93.26 |
|
3 |
KF051089 |
1151 |
4 |
KF051090 |
1164 |
97.48 |
|
3 |
KF051089 |
1151 |
5 |
KF051091 |
1167 |
96.79 |
|
3 |
KF051089 |
1151 |
6 |
KF051088 |
1216 |
97.05 |
|
3 |
KF051089 |
1151 |
7 |
KF051085 |
1128 |
96.37 |
|
4 |
KF051090 |
1164 |
5 |
KF051091 |
1167 |
98.8 |
|
4 |
KF051090 |
1164 |
6 |
KF051088 |
1216 |
98.97 |
|
4 |
KF051090 |
1164 |
7 |
KF051085 |
1128 |
98.76 |
|
5 |
KF051091 |
1167 |
6 |
KF051088 |
1216 |
98.89 |
|
5 |
KF051091 |
1167 |
7 |
KF051085 |
1128 |
98.14 |
|
6 |
KF051088 |
1216 |
7 |
KF051085 |
1128 |
98.05 |
1: KF051086; 2: KF051087; 3: KF051089; 4: KF051090; 5: KF051091; 6: KF051088; 7: KF051085
Results
DNA and 16S rDNA Gene
A total seven numbers of F. columnare strains were collected and the DNA was extracted from the strains. The extracted DNA of F. columnare was quantified spectrophotometrically and the concentration of DNA on the average was found to be between 100-200 µg/µl. A single prominent band between 1400-1500 bp fragments for 16S rDNA gene was identified in all the F. columnare isolates using universal 16S forward primer and 16S reverse primers.
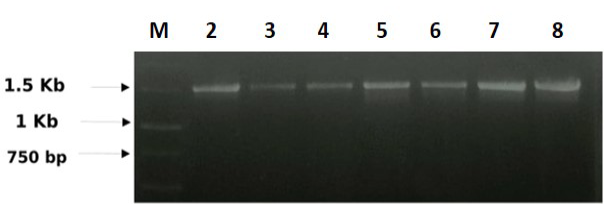
Figure 1: 16S rDNA banding pattern of F. columnare: M) 1Kb ladder (Banglore Genei, India); Lane 2) CFCCO41; Lane 3) CFCRVB43; Lane 4) CFCGFG50; Lane 5) CFCRG55; Lane 6) CFCCG62; Lane 7) CFCCSL66; Lane 8) CFCACR72
Nucleotide Sequence Submission and Phylo-genetic Tree
The seven 16S rDNA sequences obtained from the F. columnare strains were successfully sequenced and submitted in the GenBank (Figure 1). The alignment score of the seven F. columnare isolates varied from maximum of 99% to minimum of 79% (Table 2, Figure 2). The amount of variation between seven strains of F. columnare used in the present study was 1 to 21%. The maximum score was observed between KF051088 and KF051090; KF051085 and KF051090; KF051088 and KF051091. The sequence identity matrix of F. columnare isolated in our study and similar related strains isolated from the catla, rohu, goldfish, channel catfish, steelhead and ATCC (DQ005508) available at the NCBI are cited in Table 3. The phylogenetic tree was constructed based on the 16S rDNA sequences obtained in this study and other sequences available in the GenBank (Figure 2). The phylogenetic analysis was done on the basis of the bacteria isolated from infected organs as well as the whole fish. From the sequence identity matrix, it was noticed that, the identity between catla operculum KF051085 strain and catla gill KF051089 as well as between catla operculum KF051085 and catla skin lesions KF051090 were 56%. However, the identity was maximum (96%) between strains isolated from catla gill (KF051089) and catla skin lesions (KF051090). The strain of F. columnare isolated from catla available at GenBank is distantly related to our strains isolated from different organs. Similarly the strain isolated from goldfish is also distantly related to the strain (DQ005507) available in the GenBank.
Table 3: Sequence identity matrix of F. columnare strains isolated from eastern India and other sequences available at GenBank
|
Sequence |
A |
B |
C |
D |
E |
F |
G |
H |
I |
J |
K |
L |
M |
|
KF051085 |
ID |
||||||||||||
|
KF051086 |
0.261 |
ID |
|||||||||||
|
KF051087 |
0.392 |
0.243 |
ID |
||||||||||
|
KF051088 |
0.237 |
0.237 |
0.350 |
ID |
|||||||||
|
KF051089 |
0.564 |
0.252 |
0.731 |
0.222 |
ID |
||||||||
|
KF051090 |
0.563 |
0.247 |
0.747 |
0.226 |
0.963 |
ID |
|||||||
|
KF051091 |
0.249 |
0.261 |
0.247 |
0.254 |
0.258 |
0.255 |
ID |
||||||
|
DQ005507 |
0.198 |
0.193 |
0.211 |
0.182 |
0.214 |
0.220 |
0.204 |
ID |
|||||
|
JN825736 |
0.192 |
0.205 |
0.211 |
0.211 |
0.201 |
0.203 |
0.202 |
0.236 |
ID |
||||
|
AY842901 |
0.198 |
0.219 |
0.193 |
0.222 |
0.189 |
0.191 |
0.197 |
0.246 |
0.245 |
ID |
|||
|
DQ005503 |
0.191 |
0.206 |
0.205 |
0.202 |
0.199 |
0.203 |
0.206 |
0.509 |
0.248 |
0.245 |
ID |
||
|
AY842899 |
0.208 |
0.210 |
0.188 |
0.212 |
0.199 |
0.200 |
0.210 |
0.255 |
0.249 |
0.223 |
0.242 |
ID |
0.258 |
A: KF051085; B: KF051086; C: KF051087; D: KF051088; E: KF051089; F: KF051090; G: KF051091; H: DQ005507; I: JN825736; J: AY842901; K: DQ005503; L: AY842899; M: DQ005508
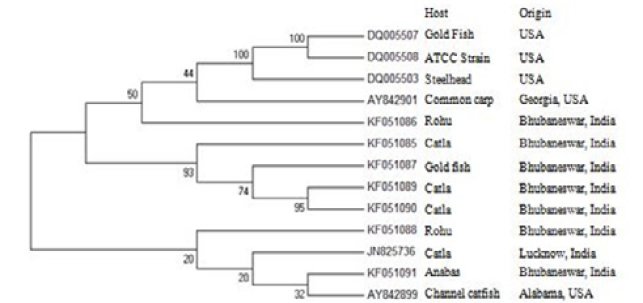
Figure 2: Phylogenetic tree based on the 16S rDNA sequences alongwith geographical origin of F. columnare
The strain isolated from anabas is closely related to the strain isolated from channel catfish with an identity of 21%. The two strains of F. columnare isolated from rohu were distantly related to each other which are placed in different clusters as revealed from phylogenetic analysis. The phylogenetic tree showed that there were at least two clades or cluster (Figure 3A and B). In the phylogenetic tree, the F. columnare sequence of goldfish (DQ005507) available at GenBank was distantly related to the F. columnare sequence of gold fish KF051087 isolated in our study. The two catla F. columnare sequences KF051089 and KF051090 used in our study were more closely related. The catla operculum KF051085 and goldfish gill KF051087 appeared before the catla gill KF051089 and catla skin lesions KF051090 in the tree, but the F. columnare sequence of catla JN825736 available at GenBank appear to be more distantly related than the catla F. Columnare sequences used in our study and remain in separate clades. The goldfish KF051087 F. columnare sequence and the ATCC strain sequence available in the GenBank appear to be more derived than the steelhead and common carp F. columnare sequences. Also, the two rohu F. columnare sequences isolated from Eastern India remain in two different clades and the rohu KF051088 F. columnare sequences appear to be more advanced than the anabas KF051091 and channel catfish AY842899 F. columnare sequences.

Figure 3A: Multiple sequence alignment of sequences obtained from 16S rRNA of F. columnare strains isolated from eastern India (KF051085, KF051086, KF051087, KF051088, KF051089, KF051090 and KF051091)
Figure 3B: Multiple sequence alignment of sequences obtained from 16S rRNA of F. columnare strains isolated from eastern India (KF051085, KF051086, KF051087, KF051088, KF051089, KF051090 and KF051091)
Discussion
F. columnare, the causal agent of columnar is disease in fishes is posing a serious problem in aquaculture industry. The use of PCR specific technique for the primary detection of fish pathogens, including F. columnare is widely debatable among microbiologists and diagnosticians in diagnostic laboratories evaluating clinical fish samples (Hiney and Smith, 1998).
The phenotypic and biochemical studies though formerly used for the species identification, have now become obsolete and are not proving useful for rapid identification and phylogenetic analysis. In the modern bacterial taxonomy the 16S rDNA gene sequence analysis is used as a confirmatory identification of different bacteria among all the cellular organisms and possess highly conserved sequence (Woose, 1987) which is used for the phylogenetic analysis between different F. columnare strains (Tiitola et al., 2006).
The present study was aimed at the molecular characterization and phylogenetic analysis of F. columnare strain isolated from different freshwater fishes of Eastern India based on the 16S rDNA gene sequences. All the strains isolated were amplified at 1.5 kbp region. The sequence analysis of these amplified product helped in the establishment of phylogenetic relationship with other F. columnare strain sequences available in the GenBank. In the phylogenetic tree, the F. columnare strain sequences isolated from same type of host were distantly related. A similar experiment by Darwish and Ismaiel (2005) with different isolates from a diseased turbot confirmed the presence of an unusual strain of F. columnare. On analysis of the phylogenetic tree it can be inferred that, the bacterial infection was not organ or species specific, however the pattern of infections varied even within the same species. The phylogenetic analysis clearly demonstrated that strains isolated from Eastern India showed variation between them along with the other strains from NCBI. It is also noticed that the strains isolated from catla were more closely related to each other indicating that the strains might have a common virulent pattern and the infection might be of systemic nature as the isolation from the external as well as the internal organs. However, our strains isolated from the different organs were distantly related to the strains available in the GenBank JN825736 (Lucknow, India). It is worthy to mention that in contrast to catla strains isolated from rohu are atypical in origin as they are distantly related indicating the nature of infection may be localized or specific to external and internal organs respectively. Further goldfish strain is closely related to strains isolated from catla. From the above it may be concluded that the strains infecting catla have the similar nature of properties to that of goldfish. However, except a few, strains isolated from the fishes belonging to the family Cyprinidae were not closely related. The bacterial strain isolated from anabas was closely related to channel catfish from Alabama, USA and distantly related to other varieties of fishes studied in the present work as well as those taken from the GenBank. Both Anabas and Channel catfish belongs to class Actinopterigii having different geographical regions. The present study revealed the strain variations among the different geographical location of the F. columnare strains; those isolated in India were distantly related to the strains of USA.
In our study, the 16S rDNA gene sequencing revealed that the strains isolated from catla and gold fish are closely related to each other whereas the strains isolated from rohu and anabas were distantly related to each other. This will help for the studying the virulence pattern, nature of infection and also for developing candidate vaccine against these different F. columnare strains.
Acknowledgments
The authors are thankful to the Director, Central Institute of Freshwater Aquaculture Bhubaneswar, India, for providing all the possible assistance and sincere co-operation for conducting this investigation.
Conflict of interest
There is no conflict of interest involved in this manuscript.
Authors’ Contribution
All the authors are equally contributed in this research work.
References





