Journal of Infection and Molecular Biology
Research Article
Plasmid Profile Analysis and Antibiotic Resistance Patterns of Shigella Isolates from Kwale County, Kenya
John Njeru1, 4*, Suleiman Saidi 2, Joseph Ngeranwa3, George Orinda3
1Centre for Microbiology Research, Kenya Medical Research Institute, P. O. Box 19464-000202 Nairobi, Kenya; 2Department of Medical Sciences, Technical University of Mombasa, P. O. Box 90420-80100, Mombasa, Kenya; 3Department of Biochemistry and Biotechnology, Kenyatta University P. O. Box 43844-00100, Nairobi, Kenya; 4Friedrich Loeffler Institute (FLI) – Institute for Bacterial Infections and Zoonosis, D-07743, Jena-Germany.
Abstract | Shigellosis caused by multidrug-resistant (MDR) Shigella strains is an important cause of morbidity and mortality in developing countries. Shigella strains usually harbour heterogeneous population of plasmids which can confer resistance to different antibiotics. A hospital based survey was conducted to determine the pattern of antimicrobial resistance and plasmid profiles in Shigella species from diarrheal patients in Kwale county. Twenty nine (8.1%) Shigella spp were obtained from 360 stool samples collected and each strain screened for antimicrobial resistance to common antimicrobial agents and presence of plasmids. High antibiotic resistance was found to Sulfamethoxazole-trimethoprim (79%), Ampicillin (75.9%) and Streptomycin (75.9%). All isolates were sensitive to ciprofloxacin, cephalosporins and Aztreonam antibiotics. Ninety seven percent of the isolates screened contained one or more plasmids ranging from 1-6. Majority were small plasmids. Species specific plasmids of 3.2kb, 9.0kb, and 3.8kb were found in Shigella flexneri, Shigella dysenteriae and Shigella sonnei respectively, while 34.5% and 79.3% MDR strains harbored large (>100kb) and middle range self-transmissible plasmids (10-100kb). In all species, no association was found between specific plasmid profile and antibiotic resistance patterns. Significant association was found between carriage of 80kb plasmid in S. dysenteriae (P<0.001), combined 48kb and 80kb plasmids in S. flexneri (P=0.002) and S. sonnei (P=0.041) and MDR phenotype. The middle range plasmids specified resistance to Sulfamethoxazole-trimethoprim, Ampicillin, Streptomycin and Tetracycline. This study reports the usefulness of plasmid profiling in detection and discrimination of Shigella strains in Kenya. The study also shows that middle range self-transferable R-plasmids are widespread among the circulating MDR Shigella strains indicating the important role of these genetic elements in the development and dissemination of multidrug resistance.
Keywords | Shigella, Antibiotic-resistance, Diarrhea, Kwale, Plasmids
Editor | Tahir Yaqub, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | July 22, 2015; Revised | August 27, 2015; Accepted | September 09, 2015; Published | October 21, 2015
*Correspondence | John Njeru, Friedrich Loeffler Institute – Institute for Bacterial Infections and Zoonosis, Jena, Germany; Email: John.Njeru@fli.bund.de
Citation | Njeru J, Saidi S, Ngeranwa J, Orinda G (2015). Plasmid profile analysis and antibiotic resistance patterns of Shigella isolates from Kwale County, Kenya. J. Inf. Mol. Biol. 3(3): 66-74.
DOI | http://dx.doi.org/10.14737/journal.jimb/2015/3.3.66.74
ISSN (Online) | 2307-5465; ISSN (Print) | 2307-5716
Copyright © 2015 Njeru et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Shigellosis is one of the most prevalent diarrheal diseases in the developing countries and a major cause of morbidity and mortality (Brooks et al., 2006; WHO, 1999; Jousilahti et al., 1997). Antibiotic resistance in Shigella species to commonly used drugs has been reported in many parts of the world (WHO, 2007). Shigellosis caused by antibiotic resistant Shigella is more frequent in African countries, reflecting the effect of widespread unregulated antimicrobial use. This has drastically reduced the antimicrobial therapeutic options and increased cost of treatment (Tesfaye et al., 2014; Mandomando et al., 2009; Gururaja et al., 2008; Huruy et al., 2008; Bogaerts et al., 1997). Previous studies in Kenya have reported multi drug resistance in Shigella strains to the commonly used antibiotics such as ampicillin, tetracycline, chloramphenicol, sulphamethoxazole/ trimethoprim, nalidixic acid and gentamicin (Sang et al., 2012; Kariuki et al., 2006; Kariuki et al., 1996). In addition, a shigellosis outbreak due to multi drug resistant Shigella dysenteriae strain was described in 1996 (Oundo et al., 1996). Despite these, detailed population based surveillance for antimicrobial resistance and molecular mechanisms involved in development and spread of resistance among enteric bacterial pathogens in Kenya is scanty.
In many parts of the world, plasmid profile analysis has been used as an epidemiological tool for investigating source of disease outbreaks, differentiating strains, and evaluating the efficacy of control measures (Ranjbar et al., 2008; Farshad et al., 2006; Nakamura et al., 1986). The antibiotic susceptibility patterns and the usefulness of plasmid profile analysis for typing Shigella strains in Kenya is poorly documented, and has not been evaluated sufficiently. This study therefore sought to determine the potential role of R-plasmids in antimicrobial resistance development and transmission in Shigella isolates obtained from diarrheic patients, and investigate whether plasmid profile analysis could be useful to discriminate between Shigella isolates for future epidemiological studies in relation to their plasmid content.
Materials and methods
Ethical Review
The approval for collection of specimens from humans was obtained from the Kenya Medical Research Institute (KEMRI) Scientific and Ethics Review Unit (SERU protocol no. 1656). Permission to conduct the study was also sought from the hospital administration. Written informed consents were obtained from all participants or guardians of minors before specimen collection.
Study Site and Population
The cross-sectional study was conducted at Kwale District hospital in Kwale County between December 2009 and August 2010. Acute diarrheal patients presenting to the outpatient clinics of the health facility were systematically assessed and consented to participate in the study. Patients who had taken antibiotics one week before recruitment were excluded. Sampling interval was based on an estimated 10 diarrheal patients treated daily at the time of study at the hospital. On the sampling day, every 2nd acute diarrheal patient meeting the case definition was enrolled into the study.
Bacteriological Analysis
Isolation and identification of pathogens were carried out using standard microbiological procedures (Ewing and Lindberg, 1984; Ewing and Edwards, 1986). Briefly, dysenteric fresh stool or rectal swab (when a subject was unable to obtain a stool sample) was obtained from consenting subjects and immediately cultured on Shigella-Salmonella agar and MacConkey agar (Difco Laboratories, BD, USA). Enrichment for Shigella was performed by culturing the specimens into selenite F broth (Oxoid, UK). All the inoculated culture media were incubated at 37°C for 24 hours. The selenite-F broth was sub cultured onto Shigella-Salmonella and MacConkey agar plates and incubated as described above. Biochemical identification of Shigella species was done using a commercial API 20E kit (Apparaeils et procedesd’ identification Montalieu vercieu, France). Briefly, a single colony was inoculated in nutrient agar (Oxoid, UK) and incubated overnight at 37°C. One to two colonies were picked from the nutrient agar plates and emulsified in sterile physiological saline to match 0.5 McFarland turbidity standards. The API cupules were then inoculated and incubated overnight at 37°C. Biochemical reactions were interpreted as instructed by the manufacturer. The Shigella strain identification was confirmed by serotyping to species level using polyvalent antisera (Denka, Seiken, Tokyo).
Antimicrobial Susceptibility Tests
The disk-diffusion method was used to test for susceptibility to 13 antibiotics commonly prescribed for treatment of shigellosis in Kenya. These included; ampicillin (Amp, 10 µg), chloramphenicol (Chl, 30 µg), streptomycin (Strp, 10µg), tetracycline (Tet, 30 µg), sulfamethoxazole-trimethoprim (Sxt, 5µg), ciprofloxacin (Cip, 5 µg), kanamycin (Kan, 30 µg), gentamycin (Gen 10 µg), ceftriazone (Ceft, 30 µg), cefixime (Cefi, 5 µg), aztreonam (Azt, 30µg), cefotaxime (Cefo, 30 µg), and nalidixic acid (Na, 30 µg). All disks were from Oxoid, UK. Briefly, log phase cultures were suspended in physiological saline to conform to 0.5 McFarland turbidity standards. Using sterile cotton tipped swabs; the organisms were aseptically spread onto Mueller-Hinton agar (Oxoid, UK) plate media. The antibiotic disks were then aseptically placed on the seeded plates. Escherichia coli ATCC 25922 was used to control for growth and disk potency. The plates were then incubated at 37°C for 24 hours. The susceptibility tests were interpreted using the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2008). The antibiograms generated were then used to cluster the isolates into various resistance profiles ranging from fully sensitive to multi drug resistant (MDR; resistant to two or more antimicrobials as described before (Mandomando et al., 2009).
Conjugation Experiments and Plasmid Profiling
Conjugation experiments were carried out using a broth mating method as described before with modifications (Dutta et al., 2002). Briefly, the Shigella strains (donors) and the Sodium azide resistant E. coli J53 (recipient) were suspended separately in sterile normal saline (0.85% NaCl) in a strength of 0.1 McFarland equivalent which is approximately 3.0 x 107 cells/mL. The donors and recipients were added together in tubes containing 5mL Mueller Hinton broth (Oxoid) in the ratios of 1:3, mixed and incubated for 8 h at 37oC without shaking. The transconjugants were recovered on MacConkey plates (Oxoid) supplemented with 200 mg/L Sodium azide (Sigma) and 30mg ampicillin (Sigma) or 30mg/L sulphamethoxazole (Sigma). The
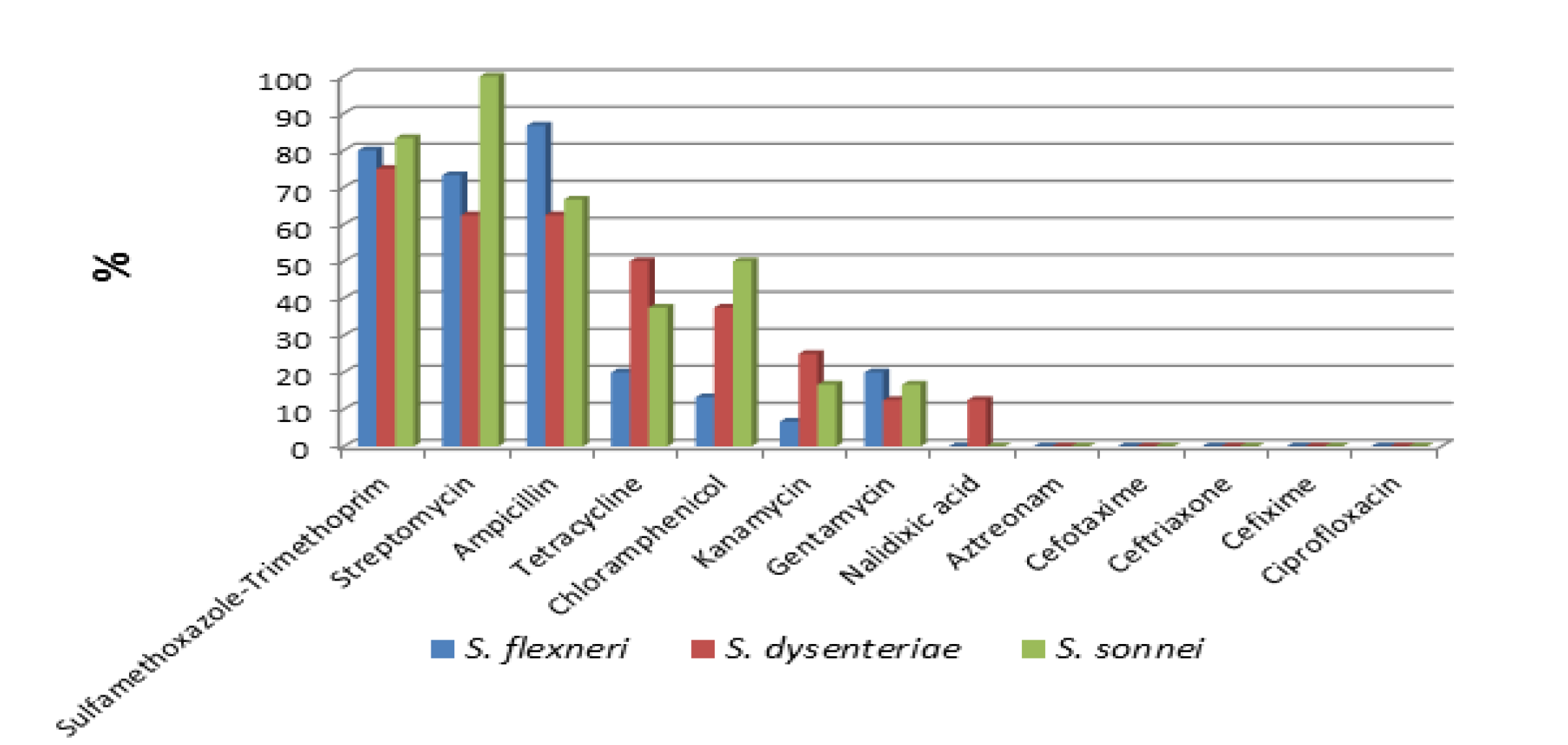
Figure 1: Percentage of antimicrobial resistance of Shigella isolates by species to specific antibiotics
susceptibility tests, analysis of resistance markers and plasmid profiling in the transconjugants was done using similar procedures as those used for donor strains.
Determination of Shigella Plasmid Profiles
Twenty nine Shigella isolates belonging to various antibiograms and transconjugants were selected and pure isolates plated on Mueller-Hinton media overnight at 37°C. The growth was used for the Plasmid DNA extraction. The extraction was performed using a commercial plasmid Mini Prep Kit (Qiagen Ltd, West Sussex, UK) according to manufacturers’ instructions. The number and molecular sizes of various plasmids isolated from representative isolates was determined by electrophoresis. Approximately 10µl of high quality plasmid DNA extracted was separated alongside plasmid DNA of Escherichia coli strain V517 as molecular size marker by electrophoresis on 1% horizontal agarose gels at 100 V for 3 hrs.
Statistical Tests
For ease of analysis, the organisms exhibiting intermediate susceptibility results were scored as resistant. The statistical associations between harbouring of resistance plasmids and multi drug resistance phenotypes were analysed by fisher exacts test. Similarly, statistical associations between the plasmid profiles and specific antibiotic resistance pattern were analysed using fisher exacts test and statistical significance considered when P value <0.05.
Results
Shigella Isolation
Overall, 29/360 (8.1%, 95%CI: 5.6-11.5) Shigella isolates were obtained. Shigella flexneri (serogroup A) accounted for 15 (51.7%), followed by S. dysenteriae (serogroup B) 8 (27.6%) and Shigella sonnei (serogroup D) 6 (20.7%). Shigella boydii (serogroup C) was not isolated in this study.
Antimicrobial Resistance
Of the 29 Shigella isolates, highest resistance was shown to sulfamethoxazole-trimethoprim (79.3%), ampicillin (75.9%) and streptomycin (75.9%), and relatively lower resistance to tetracycline (31%), chloramphenicol (27.6%), kanamycin (13.8%), and gentamycin (13.8%). Resistance to nalidixic acid was found in (3.4%) of the isolates. All the isolates were sensitive to ciprofloxacin, cephalosporins (cefotaxime, cefixime and ceftriazone) and monobactam (Aztreonam) antibiotics tested. Only one isolate was sensitive to all thirteen antibiotics tested in this study Figure 1. Resistance to one or more antibiotics was detected in 28(96.6%) of the strains, of which 25(86.2%) showed MDR characteristics. Fifteen multi drug resistance patterns were identified with sulfamethoxazole-trimethoprim, streptomycin, ampicillin and tetracycline (8 strains) and sulfamethoxazole-trimethoprim, streptomycin, ampicillin, tetracycline, chloramphenicol and gentamycin (3 strains) being the most predominant patterns Table 1.
Plasmids Characterization
Plasmid DNA analysis of the 29 Shigella isolates revealed that the isolates carried plasmids ranging from one to six. The size of the plasmids ranged from 1.5 Kb to 220Kb and majority of the plasmids were smaller than 9 Kb. Isolates containing 4 plasmids were the most abundant (10 isolates), followed by those containing 5 plasmids (7 isolates). There were five, three and two isolates harboring three, six and two plasmids respectively Table 2.
Table 1: Multi-drug resistance patterns among Shigella isolates
|
Profile |
MDR phenotype |
No. of organisms |
|
1 |
Sxt,Amp, Strp,Tet |
8 |
|
2 |
Sxt, Amp, Strp, Tet, Chl,Gen |
3 |
|
3 |
Sxt, Strp |
2 |
|
4 |
Sxt, Amp,Strp,Tet, Kan |
1 |
|
5 |
Sxt, Amp, Strp, Chl |
1 |
|
6 |
Sxt,Amp, Strp, Kan |
1 |
|
7 |
Sxt, Amp, Tet,Chl,Kan, Gen |
1 |
|
8 |
Sxt, Amp,Chl,Kan, |
1 |
|
9 |
Sxt, Amp, Strp,Tet, Chl |
1 |
|
10 |
Sxt, Amp,Na,Tet,Chl |
1 |
|
11 |
Sxt, Amp, Tet |
1 |
|
12 |
Sxt,Gen |
1 |
|
13 |
Amp,Strp |
1 |
|
14 |
Amp,Strp,Gen |
1 |
|
15 |
Amp,Chl,Kan,Tet,Gen |
1 |
|
Total |
25 |
MDR (multi drug resistance); Ampicillin (Amp, 10 µg); Chloramphenicol (Chl, 30 µg); Streptomycin (Strp, 10µg); Tetracycline (Tet, 30 µg); Sulfamethoxazole-Trimethoprim (Sxt, 5µg); Kanamycin (Kan, 30 µg); Gentamycin (Gen 10 µg); Kanamycin (Kan, 30 µg)
Table 2: Distribution of plasmids among Shigella isolates
|
No. of plasmids |
S. flexneri |
S. dysenteriae |
S. sonnei |
No. of isolates |
|
0 |
1 |
0 |
0 |
1 |
|
1 |
0 |
1 |
0 |
1 |
|
2 |
1 |
0 |
1 |
2 |
|
3 |
1 |
2 |
2 |
5 |
|
4 |
5 |
4 |
1 |
10 |
|
5 |
5 |
1 |
1 |
7 |
|
6 |
2 |
0 |
1 |
3 |
|
Total |
15 |
8 |
6 |
29 |
Unlike fifteen profiles distinguished by the antimicrobial susceptibility testing, plasmid profiling of the Shigella species distinguished nineteen different plasmid patterns. Large (>100kb), middle range (10-100kb) and small (<10kb) plasmids were demonstrated by the isolates. The plasmid patterns were distinct for each Shigella spp, but 1.5kb and 2.0kb plasmids were shared among the three Shigella species occurring in 75.9% and 37.9% of the isolates respectively. In addition 3.2kb, 3.8kb and 9.0kb species specific plasmids were exclusively present in S. flexneri, S. sonnei and S. dysenteriae respectively.
Plasmid Profiles and Association with Antibiotic Resistance in S. flexneri
Eight plasmid groups containing 2 to 6 plasmids of between 1.5-220kb were identified in S. flexneri isolates. The large plasmid(220kb) was found in 28.6% isolates while 3.2kb was the most abundant among small plasmids occurring in 13 (93%) of the isolates. Other small plasmids (1.5kb and 4.1 kb) occurred in 78.6% and 50% of the isolates respectively. The small range plasmids of sizes 2.0kb, 2.8kb and 7.0kb were present in very small proportions. The middle range plasmids (48kb and 80kb) were each present in 78.6% and 28.6% of the S. flexneri isolates respectively Table 3. Statistical analysis did not reveal association between plasmid profiles and specific antibiotic resistance pattern in S. flexneri. All isolates with multi-drug resistance characteristics harbored at least one of the middle range plasmids (48kb and/or 80kb) while none of the isolates lacking MDR phenotype harbored any of these plasmids. A statistically significant association was demonstrated between presence of 48kb and 80kb plasmids and multi drug resistance phenotype among in this serogroup (Fishers Exact Test, p=0.002).
Plasmid Profiles and Association with Antibiotic Resistance in S. dysenteriae
Six plasmid groups containing 1 to 5 plasmids of between 1.5-220kb were identified in S. dysenteriae isolates. The large plasmid of 220kb was found in 3 of 8 (37.5%) isolates and small size plasmid (9.0kb) was detected in all but one isolate. The common small plasmids (1.5 and 2.0kb) occurred in 62.5% and 50% of the isolates respectively. Small plasmids (4.1kb) and (5.2kb) were less common among the isolates. The middle range plasmid (48kb) was absent in this serogroup, but 80kb plasmid was found in 5 of 8 (62.5%) isolates Table 3. A statistically significant association was found between carriage of 80kb middle range plasmid and MDR phenotype among S. dysenteriae (Fishers Exact Test, p<0.001).
Plasmid Profiles and Association with Antibiotic Resistance in S. sonnei
Five plasmid groups containing 2 to 6 plasmids of between 1.5-180kb were identified in S. sonnei isolates. The large plasmid of 180kb was found in 3 of 6(50%) isolates. Among small size plasmids, 1.5kb plasmid was found in all the S. sonnei isolates while 3.8kb and 2.0kb sizes occurred in 66.7% and 50% of the isolates. The middle range plasmids (48kb and 80kb) were each present in 66.7% and 50% of the S. sonnei isolates respectively Table 3. As in other species, analysis did not reveal any association between plasmid profiles and specific antibiotic resistance pattern in this serogroup. However four of six isolates with multi-drug resistance harboured at least one of the middle range plasmids (48kb and/or 80kb), while two MDR strains lacked
Table 3: Plasmid profiles in different Shigella species
|
Plasmid(Kb) |
220 |
180 |
80 |
48 |
9.0 |
7.0 |
5.2 |
4.1 |
3.8 |
3.2 |
2.8 |
2.0 |
1.5 |
No. of organisms |
|
S. flexneri |
||||||||||||||
|
Profile 1 |
+ |
+ |
+ |
+ |
+ |
+ |
2 |
|||||||
|
Profile 2 |
+ |
+ |
+ |
+ |
2 |
|||||||||
|
Profile 3 |
+ |
+ |
+ |
+ |
5 |
|||||||||
|
Profile 4 |
+ |
+ |
+ |
+ |
+ |
1 |
||||||||
|
Profile 5 |
+ |
+ |
+ |
1 |
||||||||||
|
Profile 6 |
+ |
+ |
1 |
|||||||||||
|
Profile 7 |
+ |
+ |
+ |
+ |
1 |
|||||||||
|
Profile 8 |
+ |
+ |
+ |
+ |
1 |
|||||||||
|
Total |
14 |
|||||||||||||
|
S. dysenteriae |
||||||||||||||
|
Profile 1 |
+ |
+ |
+ |
+ |
+ |
1 |
||||||||
|
Profile 2 |
+ |
+ |
+ |
2 |
||||||||||
|
Profile 3 |
+ |
+ |
+ |
+ |
2 |
|||||||||
|
Profile 4 |
+ |
+ |
+ |
+ |
1 |
|||||||||
|
Profile 5 |
+ |
1 |
||||||||||||
|
Profile 6 |
+ |
+ |
+ |
+ |
1 |
|||||||||
|
Total |
8 |
|||||||||||||
|
S. sonnei |
||||||||||||||
|
Profile 1 |
+ |
+ |
+ |
+ |
+ |
+ |
1 |
|||||||
|
Profile 2 |
+ |
+ |
+ |
2 |
||||||||||
|
Profile 3 |
+ |
+ |
+ |
+ |
1 |
|||||||||
|
Profile 4 |
+ |
+ |
1 |
|||||||||||
|
Profile 5 |
+ |
+ |
+ |
+ |
+ |
1 |
||||||||
|
Total |
6 |
|||||||||||||
+ (Plasmid present)
any of these plasmids. A statistically significant association was demonstrated between harbouring of 48kb and 80kb plasmids and multi drug resistance phenotype among S. sonnei. (Fishers Exact Test, p=0.041).
Plasmids Conjugation and Analysis for Transferable Antibiotic Resistance
Twenty two MDR isolates were selected for conjugation experiments, comprising of 10 S. flexneri, 6 S. sonnei and 6 S. dysenteriae. Resistance phenotypes were only transferred in 32% of isolates either in full or partly to Sodium azide resistant E. coli J53 strain when they were co cultivated. These included those that were resistant to six antibiotics Table 4. Analysis of the transconjugants arising from conjugation with S. flexneri isolate (Sh27) that exhibited ampicillin resistance (AmpR) and streptomycin resistance (StrpR) revealed that the resistance elements were located on a self-transferable 80kb plasmid. This was characterized by transfer of its full antibiotic resistance profile (AmpR) and (StrpR) to E. coli J53 strain. Two other S. flexneri isolates; (Sh01) and (Sh12) that showed SxtR, AmpR, StrpR, TetR, KanR and SxtR, AmpR, StrpR, TetR respectively, each conjugally transferred two R-plasmids (48kb and 80kb). While the latter transferred its full spectra of resistance gene cassettes, the former transferred the resistance gene cassettes but not the one conferring resistance to kanamycin. This indicated that all but kanamycin resistance genes were possibly located on these self-transferable middle range plasmids Table 5. A plasmid of 80kb was conjugally transferred by S. dysenteriae (Sh08) carrying AmpR StrpR ChlR KanR TetR GenR resistance genes to E coli J53 strain. Analysis of the transconjugants revealed that only ampicillin and streptomycin resistance phenotypes were transferrable Table 5.
One S. sonnei isolate (Sh09) showing SxtRAmpR StrpR TetR ChlR conjugally transferred two plasmids of 80kb and 48kb accompanied by transfer of its resistance profile but not resistance to chloramphenicol. Additionally, another S. sonnei isolate (Sh17) of SxtRAmpR StrpR TetR ChlR GenR phenotype harboring six plasmids, conjugally transferred two middle range plasmids 80kb and 48kb as well as the
Table 4: Antibiotic resistance phenotypes transferred from MDR Shigella to recipient E. coli J53 strain
|
Isolate |
Identity |
Donor profiles |
Tn |
Transconjugants profiles |
|
Sh17 |
SS |
SxtRAmpR StrpR TetR ChlR GenR |
+ |
SxtRAmpR StrpR TetR ChlR |
|
Sh05 |
SS |
SxtRAmpR StrpR TetR ChlR GenR |
+ |
SxtRAmpR StrpR TetR |
|
Sh09 |
SS |
SxtRAmpR StrpR TetR ChlR |
+ |
SxtRAmpR StrpR TetR |
|
Sh01 |
SF |
SxtRAmpR StrpR TetR KanR |
+ |
SxtRAmpR StrpR TetR |
|
Sh12 |
SF |
SxtRAmpR StrpR TetR |
+ |
SxtRAmpR StrpR TetR |
|
Sh27 |
SF |
AmpR StrpR |
+ |
AmpR StrpR |
|
Sh08 |
SD |
AmpR StrpR ChlR KanR TetR GenR |
+ |
AmpR StrpR |
|
Sh10 |
SF |
SxtRAmpR StrpR TetR |
- |
- |
|
Sh06 |
SS |
SxtRAmpR StrpR GenR |
- |
- |
|
Sh07 |
SF |
SxtRAmpR StrpR TetR |
- |
- |
|
Sh15 |
SS |
SxtRAmpR StrpR KanR |
- |
- |
|
Sh04 |
SD |
SxtRAmpR ChlR TetR KanR GenR |
- |
- |
|
Sh13 |
SF |
SxtRAmpR ChlR KanR |
- |
- |
|
Sh20 |
SD |
SxtRAmpR TetR ChlR |
- |
- |
|
Sh11 |
SF |
SxtRAmpR StrpR TetR |
- |
- |
|
Sh14 |
SF |
SxtRAmpR StrpR TetR ChlR GenR |
- |
- |
|
Sh25 |
SS |
SxtRAmpR StrpR TetR |
- |
- |
|
Sh19 |
SF |
SxtRAmpR StrpR |
- |
- |
|
Sh28 |
SD |
SxtRAmpR TetR |
- |
- |
|
Sh02 |
SD |
SxtRAmpR StrpR TetR |
- |
- |
|
Sh22 |
SD |
SxtRAmpR TetR ChlR NaR |
- |
- |
|
Sh03 |
SF |
AmpR StrpR GenR |
- |
- |
SD (S. dysenteriae); SS (S. sonnei); SF (S. flexneri); Tn (Transconjugant); Ampicillin resistance (AmpR, 10 µg); Chloramphenicol resistance (ChlR, 30 µg); Streptomycin resistance (StrpR, 10µg); Tetracycline resistance (TetR, 30 µg); Sulfamethoxazole-Trimethoprim resistance (SxtR, 5µg); Kanamycin resistance (KanR, 30 µg); Gentamycin resistance (GenR 10 µg); Kanamycin resistance (KanR, 30 µg)
Table 5: Antibiotic resistance profiles and the associated R-plasmids transferred from MDR Shigella to recipient E. coli J53 strain
|
Isolate |
Identity |
Donor profile |
Donor plasmids |
Tn profiles |
Transconjugant plasmids(Mw) |
|
Sh17 |
SS |
SxtRAmpRStrpRTetRChlRGenR |
6 |
SxtRAmpRStrpR TetR ChlR |
3(180KB,80kb,48kb) |
|
Sh05 |
SS |
SxtRAmpRStrpR TetR ChlR GenR |
5 |
SxtRAmpRStrpR TetR |
2(80kb,48kb) |
|
Sh09 |
SS |
SxtRAmpRStrpRTetR ChlR |
3 |
SxtRAmpRStrpR TetR |
2(80kb,48kb) |
|
Sh01 |
SF |
SxtRAmpR StrpRTetRKanR |
6 |
SxtRAmpRStrpR TetR |
2(80kb,48kb) |
|
Sh12 |
SF |
SxtRAmpR StrpR TetR |
4 |
SxtRAmpRStrpR TetR |
2(80kb,48kb) |
|
Sh27 |
SF |
AmpR StrpR |
2 |
AmpR StrpR |
1(80kb) |
|
Sh08 |
SD |
AmpR StrpR ChlR KanR TetR GenR |
5 |
AmpR StrpR |
1(80kb) |
SD (S. dysenteriae); SS (S. sonnei); SF (S. flexneri); Tn (Transconjugant); Ampicillin resistance (AmpR, 10 µg); Chloramphenicol resistance (ChlR, 30 µg); Streptomycin resistance (StrpR, 10µg); Tetracycline resistance (TetR, 30 µg); Sulfamethoxazole-Trimethoprim resistance (SxtR, 5µg); Kanamycin resistance (KanR, 30 µg); Gentamycin resistance (GenR 10 µg); Kanamycin resistance (KanR, 30 µg)
large plasmid of 180kb. All but gentamycin resistance gene cassettes were transferred in this isolate. The third S. sonnei isolate (Sh05) with the same resistance profile (SxtRAmpR StrpR TetR ChlR GenR) conjugally transferred only the two middle range plasmids 80kb and 48kb. Similarly the resistance gene cassettes transferred in this isolate were those conferring resistance to SxtRAmpR StrpR TetR but resistance to chloramphenicol and gentamycin were nontransferable (Table 5).
It was also showed that resistance to gentamycin and kanamycin were not transferable traits among the MDR Shigella isolates while the resistance to chloramphenicol was associated with the transfer of large plasmid (180kb) in S. sonnei.
Discussion
In the present study, plasmid profiling distinguished more Shigella strains than did antimicrobial susceptibility patterns. This suggests the presence of different endemic strains or a high rate of horizontal genetic instability amongst the circulating strains or both. Plasmid profiling of Shigella strains may therefore be useful in future epidemiological studies in the Kenyan Coast. This finding agrees with the studies by Tacket and Cohen (1983) in Bangladesh, Olukoya and Oni (1990) in Nigeria and Ranjbar et al. (2008) in Iran who found plasmid profiling more discriminative than antimicrobial susceptibility testing in distinguishing Shigella strains. Only ten of the 29 isolates (34.5%) were found to harbour large plasmids (180kb and 220kb). These plasmids are associated with invasiveness properties of Shigella strains (Sansonnetti et al., 1982; Casalino et al., 1994; Farshad et al., 2006). The absence of these plasmids in most of the Shigella species suggest that these plasmids were possibly lost during storage and subsequent subculture of the isolates. This has been documented before by authors of similar studies (Kopecko et al., 1980; Lin and Chang, 1992; Bebora et al., 1994).
In this study, most plasmids ranged from 1 to 2kb, 3 to 4kb and 5 to 9 kb. These were detected in 86.9%, 78.3% and 47.5% of all isolates, respectively, irrespective of their antibiotic susceptibility phenotypes. In particular, 3.2kb, 9.0kb and 3.8kb small plasmids were exclusively present in S. flexneri, S. dysenteriae and S. sonnei, respectively. This strongly suggests that these are core Shigella plasmids that constitute a stable gene pool and that can be used as preferred markers for identification of each of the corresponding serogroup in future epidemiological studies. Small plasmids of similar sizes were demonstrated in the previous studies in Malasyia (Hoe et al., 2005), in India (Dutta et al., 2002), in Peru (Fernandez-Prada et al., 2004) and in Iran (Shohreh et al., 2006). Significant statistical association was found between presence of middle range plasmids (48kb and 80kb) and MDR phenotype among the Shigella isolates. This indicates that these plasmids harboured the gene cassettes that confer resistance to the commonly used antibiotics. This corresponds well with similar reports by Olukoya et al. (1990) in Nigeria and Ling et al. (1993) in England.
The study showed that middle-sized self-transferable plasmids (48kb and 80kb) were simultaneously or singly harboured in 79.3% and transferred by 32% of the MDR Shigella isolates, to the recipient J53 strain. These plasmids were found to be responsible for the transfer of resistance gene cassettes conferring resistance to the four affordable and most commonly used antibiotics for treatment of shigellosis (ampicillin, sulfamethoxazole-trimethoprim, streptomycin and tetracycline) in Kenya. Specifically, transfer of ampicillin and streptomycin resistance was significantly associated with a single plasmid of 80kb while transfer of combined resistance to ampicillin, sulfamethoxazole-trimethoprim, streptomycin and tetracycline was associated with joint transfer of 48kb and the 80kb plasmids. These results suggest that the genes conferring resistance to ampicillin and streptomycin are located on the 80kb plasmid while those conferring resistance to sulfamethoxazole-trimethoprim and tetracycline are located on 48kb plasmid. It is also possible that the resistance genes to ampicillin and streptomycin are present in the two 48 kb and 80 kb plasmids. A similar study by Seol et al. (2006) on S. sonnei in South Korea documented a 70–80kb R-plasmid that was responsible for transfer of sulfamethoxazole-trimethoprim, streptomycin and tetracycline resistance. However, the findings of this study do not correspond to the findings by Fernandez-Prada et al. (2004) in Peru. Their study did not find a correlation between presence of a particular plasmid and antibiotic-resistance profile in S. flexneri. Considering that the R-plasmids harbouring genes encoding MDR were transferable en masse or partly, acquisition of a large number of drug resistance genes by human enteric pathogens in a single step is very likely. The transfer of large plasmid (180 kb) by S. sonnei and subsequent acquisition of resistance to chloramphenicol shows that the genes conferring resistance to this drug are highly likely located on this plasmid. It was observed in this study that despite several MDR donors harbouring kanamycin and gentamycin resistance genes, the resistance phonotypes were not transferrable by conjugation indicating that these genes were most likely chromosomally located. On the other hand, the genes could possibly be located on plasmids that are not conjugative, but further research is worth to definitively demonstrate this hypothesis. In general, plasmid transfer via conjugation has been documented as an active mechanism for rapid DNA exchange or dissemination of antimicrobial resistance in bacterial populations (Gregory et al., 2008). This mechanism may ultimately accelerate the evolution of Shigella pathotypes contributing towards the plasticity of their genomes and the diverse combinations of resistance factors.
Conclusion
This is the first study to describe the potential usefulness of plasmid profiling in detection and discrimination of Shigella strains in Kenya. Our study shows that middle range, self-transferable R- plasmids carrying gene cassettes which confer resistance to different classes of antimicrobials, are widespread among MDR Shigella strains in the Kenyan coast. This together with the evidence of co transfer of multiple resistance genes indicates the important role of these genetic elements play, in the development and dissemination of multidrug resistance. This would further limit the use of the cheap and readily available classes of antibiotics used in empiric treatment of Shigellosis and even compromise the eventual use of third line antimicrobials. Further research involving nucleotide sequencing, screening and detection of the specific localization of gene cassettes encoding resistance to a range of available antibiotics is highly recommendable.
Acknowledgements
The authors would like to thank Prof. Sam Kariuki, Director of Centre for Microbiology Research-KEMRI for providing facilities to carry out this study. We are also grateful to Dr. John Kiiru for the scientific guidance and support. We also thank the Director Kenya Medical Research Institute (KEMRI) for the financial support to conduct the study. We acknowledge the administration, clinicians and laboratory staff of the Kwale hospital and CMR laboratory for the technical support.
Competing Interests
The authors declare that there exist no competing interests.
AUTHORS CONTRIBUTION
JNM was involved in conception, design, data collection, data analysis and drafting the manuscript. SS was involved in conception, design, data collection and coordination of the study. JJN and OG were presented concept, coordinated study and analysed the data. All authors participated in writing of the manuscript and approved submission of the final manuscript.
References





