Journal of Infection and Molecular Biology
Short Communication
Biological and Molecular Characterization of Newcastle Disease Virus through Haemagglutinin-neuraminidase Gene Isolated from Lahore District
Hashaam Habib1, Shafqat Fatima Rehmani2, Nadia Mukhtar1, Tasra Bibi1, Abdul Wajid2, 3
1Department of Microbiology; 2Quality Operations Laboratory and 3Institute of Biochemistry and Biotechnology, University of Veterinary and Animal Sciences, Lahore, 54600, Pakistan.
Abstract | Newcastle disease (ND) is a fatal and contagious viral disease of many avian species predominantly domestic poultry that creates a constant threat to the poultry industry around the world including Pakistan. The efficient diagnosis of NDV strains become a challenge due to complex clinico-pathological picture and high genetic variability. In this study, one hundred ND suspected samples were collected from the proximity of Lahore district during 2010. Upon virus issolation, these samples were subjected to haemagglutination (HA) and haemagglutination inhibition (HI) tests for the confirmation of NDV. Ten confirmed samples were pathogenic as determined by ICPI and MDT. Five NDV isolates representing different geographical areas of Lahore were subjected for the amplification of full length hemagglutinin-neuraminidase (HN) gene (1892 bp). Subsequently, a partial HN (519 bp) was sequenced to characterized the NDV isolates. All the isolates had closed phylogenetic relationship with previously characterized Pakistani and Indonesian isolates within genotype VII. The present investigation provides essential information on the genetic nature of HN protein of NDV circulating in Pakistan and emphasizes the importance of study on disease diagnosis and control.
Keywords | Newcastle disease, ICPI, MDT, HN gene, Phylogenetic analysis
Editor | Tahir Yaqub, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | September 15, 2014; Revised | January 06, 2015; Accepted | January 13, 2015; Published | January 26, 2015
*Correspondence | Shafqat Fatima Rehmani, University of Veterinary and Animal Sciences, Lahore, Pakistan; Email: rehmani.shafqat@uvas.edu.pk
Citation | Habib H, Rehmani SF, Mukhtar N, Bibi T, Wajid A (2015). Biological and molecular characterization of Newcastle disease virus through haemagglutinin-neuraminidase gene isolated from Lahore district. J. Inf. Mol. Biol. 3(2): 28-33.
DOI | http://dx.doi.org/10.14737/journal.jimb/2015/3.2.28.33
ISSN (Online) | 2307-5465; ISSN (Print) | 2307-5716
Copyright © 2015 Habib et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Newcastle disease (ND) is one of the most important viral diseases of birds affecting domestic poultry, caged pet birds and wild birds throughout the world. The poultry sector is one of the most organized and vibrant segment of the agriculture industry of Pakistan and generates direct and indirect employment. This sector generates direct and indirect employment and income for more than 1.5 million people. Its contribution in agriculture and livestock is 6.4% and 11.5% respectively. However, this industry is always threatened by various viral and bacterial diseases among which ND is on the top. The disease is complicated due to different pathotypes and strains of the virus that may induce enormous variation in the severity of disease characterized by fatal respiratory and neurological pathogenesis. During the recent years, from 2009 to date, a number of vNDVs have been isolated from commercial, rural and wild birds in Pakistan (Khan et al., 2010; Munir et al., 2012a; Munir et al., 2012b; Shabbir et al., 2012). Natural routes of infection (nasal, oral, and ocular) appear to emphasize the respiratory nature of the disease, while intramuscular, intravenous and intracerebral routes appear to enhance the neurologic signs.
The NDV belongs to family paramyxoviridae, sub-family paramyxovirinae in the genus of Avulavirus (Mayo, 2002). The paramyxoviridae encompasses a diverse group of viruses consist of a negative sense single-stranded, non-segmented RNA molecule. Nucleotide sequencing of the NDV genome has encoding six proteins including hemagglutinin-neuraminidase protein (HN), fusion protein (F), necleocapsid protein (NP), matrix protein (M), phosphoprotein (P), and RNA-directed RNA polymerase (L) (Alexander, 2003). The viruses consist of an outer lipoprotein membrane composed of lipid bilayer and three membranes bound proteins: Among these, the hemagglutinin-neuraminidase (HN) and Fusion (F) proteins are two transmembranal glycoproteins and an inner non-glycosylated matrix protein (M) (Smith et al., 2009). The two transmembrane glycoproteins, the fusion protein (F) and virus attachment protein the HN, form spike like protrusions on the outer surface of the viruses. The HN glycoprotein is a type II integral membrane and multifunctional glycoprotein and is the key antigenic determinant of paramyxoviruses, including NDV, parain fluenza virus 5 (PIV5), mumps virus, sendai virus and human paraunfluenza viruses 1-4 (hPIV1-4) (Wang et al., 2013; Lamb et al., 1996). HN glycoprotein is responsible for the attachment of the viruses to sialic acid containing receptors in the plasma membrane of the host cells (Chaturvedi et al., 2011). Moreover, its neuroaminidase activity hydrolyses the sialic acid containing molecules and likely to release the new-born viral particles from the target cells. The hemagglutinin-neuroaminidase (HN) protein of NDV plays a significant role in virus virulence and tissue tropism. However, it promotes the fusion activity of F protein, thus allowing the virion to penetrate the cell surface.
The severity of the disease depends on the virus strains and the host species (Yuan et al., 2012). NDV isolates are categorized into highly pathogenic (velogenic), intermediate (mesogenic) and low (lentogenic) strains. Both HN and F play significant role to initiate the infection (Romer-Oberdorfer et al 2003; Huang et al., 2004). However, the HN proteins show high sequence similarity from different paramyxoviruses, generally in the core neuraminidase (NA) domain. The hemagglutinin-neuroaminidase (HN) protein of NDV strains having different amino acids length: 571 aa, (Munir et al., 2012b; Wang et al., 2013) 577 aa (Ponnusamy et al., 2009) (present study), 580 aa (Huang et al., 2004), 581 aa, (Tan et al., 1995) 585 aa (Hu et al., 2010) and 616 aa (Yuan et al., 2012). Sequence analysis of HN gene revealed that many low virulent enteric NDV strains have a large open reading frame (ORF) (616 amino acid, aa) with additional 45 aa at its C-terminus when compared with that of some virulent and certain less virulent NDV strains (571 and 577 aa). Avirulent NDV strains e.g. D26, Ulster and Queensland having precursor HNo of 616 aa residues necessitate a post translational cleavage to create a biologically active HN protein.
To obtain a better understanding of the genetic relationships of the NDV isolates, genomic sequence analysis of the region covering the cleavage site of the F protein and C-terminal extension of the HN gene would be helpful for better intervention strategies.
One hundred ND suspected samples were collected from different sources at the vicinity of district Lahore, Punjab province Pakistan during 2009-10. Buccal and cloacal swabs from live birds along with various organs like trachea and spleen from morbid/dead birds were collected. A 0.2 ml aliquot of organ/swab suspensions were inoculated separately into the allantoic cavity of 9 day old specific pathogen-free (SPF) embryonated hen’s eggs for virus isolation. The allantoic fluid was collected after 3-day incubation at 37°C and tested with haemagglutination assay (HA). Positive samples were confirmed as PMV-1 with haemagglutination inhibition (HI) according to the procedure described by Alexander and Chettle (1977).
The pathogenic potential of ten isolates was evaluated using standard assay methods to determine the intracerebral pathogenicity index (ICPI) in 1-day-old chickens and mean death time (MDT) was determine by inoculating the allantoic cavities of 9-11 days old embryo according to international OIE standards (Alexander, 2008).
Two hundred and fifty μL infected allantoic fluid was used for RNA extraction using TriZol® LS Reagent (Invitrogen, Carsbad, CA, USA) in accordance with the manufacturer’s instructions. The extracted RNA pellet was re-suspended in 50 μL nuclease free water (Promega, USA), and kept in -20ºC for later use. The extracted viral genomic RNA was used for cDNA using cDNA synthesis kit (Verso™ cDNA synthesis kit by thermo scientific) as recommended by the manufacturer. The thermo profile for the reverse transcription was 42°C for 30 min and 95°C for 2 min. The cDNA was stored at –20°C until further use. In order to obtain the complete HN gene sequence, the PCR amplification was performed using the previously reported primers F (NDV5HN) 5’-GTAGGCTAGCAAGAGAGGCCGCCCCTCAAT-3’ and R (NDV3HN) 5’-CGAGCCCGGGCCGGCATTCGGTT TGATTCTTG-3’ (Peeters et al., 2001). The thermo profile used was; pre-denaturation at 94°C for 5 min and then 35 cycles of denaturation at 94°C for 1 min, Annealing at 62° C for 45 sec, Extension at 72°C for 2 min and Final extension at 72°C for 7 min. Then internal primers were designed for the amplification of a short (519bp) product of HN gene to be sequenced. The primers used were NDV-F 5’-CATACACAACATCAACATG-3’ and NDV-R 5’-GGTAGCCCAGTTAATTTCCA-3’. The thermo profile used was pre-denaturation at 94°C for 5 min and then 35 cycles of denaturation at 94°C for 30 Sec, Annealing at 54° C for 30 Sec, Extension at 72°C for 1 min and Final extension at 72°C for 10 min.
The PCR amplicons were subjected for DNA sequencing using ABI 3130 automated sequencer (Applied Biosystem Inc, Foster City, CA) at Quality Operations Laboratory (QOL), University of Veterinary and Animal Sciences, Lahore, Pakistan. Nucleotide sequence assembling, editing and analyses were conducted by using Codon Code Aligner (v 4.1.1 Codon Code Corporation). Multiple alignments and Phylogenetic analysis of the partial sequences of HN gene along with corresponding sequences from the GenBank were performed using ClustalW multiple alignment algorithm in MEGA5 (Tamura et al., 2011). The evolutionary history was inferred by using Neighbour Joining with Kimura two-parameter model along with 1,000 bootstraps value.
In this study, 100 ND suspected samples were collected from chickens at the vicinity of Lahore district of Punjab province were characterized biologically. Haemagglutination is considered as a characteristic feature of paramyxoviruses and orthomyxoviruses. After cultivation of virus in embryonated eggs, Spot HA positive allanto-amniotic fluid (AAF) were collected and subjected to hemagglutination (HA) and hemagglutination inhibition (HI) tests for the confirmation of ND and Avian influenza viruses. No influenza virus A was detected and the NDV isolation rate was higher from cloacal swabs as compared to buccal swabs. Similarly the NDV was isolated at a higher rate from trachea as compared to spleen samples. In earlier studies breeding ducks were selected for the isolation of various viruses. Those isolates were found positive for HA tests and all of them were detected as NDV on the bases of HI test and virus neutralization assays performed using antisera against AIV and NDV.
Table 1 represent ten NDV isolates recovered from clinically diseased chickens from the vicinity of district Lahore, Punjab, Pakistan. The virulence assay of the infected embryos revealed the MDT ranged from 40 to 104 h, classified the NDV isolates into velogenic, mesogenic and lentogenic (<60h: velogenic, 60-90h: mesogenic and >90h: lentogenic). The ICPI values ranged from 0.41 to 1.65, biologically characterized the isolates into three various form of NDV. The intracloacal inoculation of chickens revealed that the high virulent strains produced viscerotropic lesions while the low virulent isolates produced neurotropic lesions in chickens.
Table 1: Biological and molecular characteristics of NDV isolates recovered from chickens in Pakistan during 2010
|
|
|
|
|
Pathogenicity |
|
|
|
S.NO |
Year of collection |
Host |
ICPI |
MDT |
HN extension |
|
|
1 |
Chicken/QOL/04/10 |
2010 |
Chicken |
0.70 |
56 |
571 |
|
2 |
Chicken/QOL/06/10 |
2010 |
Chicken |
0.70 |
60 |
571 |
|
3 |
Chicken/QOL/33/10 |
2010 |
Chicken |
1.27 |
60 |
571 |
|
4 |
Chicken/QOL/38/10 |
2010 |
Chicken |
1.26 |
104 |
- |
|
5 |
Chicken/QOL/41/10 |
2010 |
Chicken |
1.41 |
64 |
- |
|
6 |
Chicken/QOL/53/10 |
2010 |
Chicken |
1.65 |
48 |
571 |
|
7 |
Chicken/QOL/55/10 |
2010 |
Chicken |
1.63 |
40 |
571 |
|
8 |
Chicken/QOL/58/10 |
2010 |
Chicken |
1.27 |
64 |
- |
|
9 |
Chicken/QOL/68/10 |
2010 |
Chicken |
0.59 |
96 |
- |
|
10 |
Chicken/QOL/81/10 |
2010 |
Chicken |
0.41 |
96 |
- |

Figure 1: Sequence alignment of C-terminus of HN gene of various NDV strains, the sequences of the current study, LaSota (JF950510), Ulster (M19478), R2B (JX316216) and previously characterized PAK strains Chicken/BYP/Lahore/2010 (JN682200). The C-terminus amino acids are colored green
Nucleotide sequencing and subsequent deduction of the amino acid sequence covering the C-terminus of the HN protein showed 571 amino acids in Pakistani NDV strains (Figure 1). Furthermore, we discovered that the previous characterized Pakistani NDV isolates examined had an HN protein consisting of 571 amino acids (Munir et al., 2012b). The size of the HN protein of NDV strains is highly variable due to the position of the stop codon, which give rise to different sizes of the predicted protein product. The HN protein of the NDV strains has different amino acid lengths e.g. 571aa, 577aa, 581aa and 616aa (Romer-Oberdorfer et al 2003; Yuan et al., 2012). All the characterized NDV isolates have HN protein of 571 amino acids (Munir et al., 2012b), which is a feature of virulent NDV strains (Maminiaina et al., 2010). Most NDV virulent and less virulent strains exhibit HN proteins of 571 and 577 aa, since termination codons are located before the HNo stop codon. The high virulent NDV strains were reported to have 571 aa residues while length of 577 aa residues were reported in some virulent and less virulent strains like Clone-30 (Romer-Oberdorfer et al 2003). The 616 amino acids the largest CDS of HN gene has been in avirulent NDV strains like Ulster 2C, Queensland V4, D26 was detected as precursor protein HNo (Yuan et al., 2012). The precursor protein HNo having 45 aa residues proteolytically remove as a small glycosylated fragment from the C-terminus to form biologically active glycoprotein HN protein (Hooper et al., 1999). It has been demonstrated that the NDV Ulster protein extra C-terminal 45 amino acids have residues within that blocks two key receptor binding regions necessary for attachment to cells and virus entry into host. This unique evolutionary adaptation has been consistent with a significant role in modulating NDV pathogenicity. The primary site of interest in disease potential of NDV is the Fo cleavage signal of an isolate (Czegle´di et al., 2006). However, the HN extension site has been suggested is a predictive marker for the NDV lineages. The two surface glycoproteins the fusion (F) and haemagglutinin-neuraminidase protein, activate NDV entry into cells and variations in both proteins are linked to differences in strain specific virulence.
Phylogenetic analysis was conducted using the nucleotide sequence of five NDV isolates along with the sequences of different NDV isolates available in GenBank. From the topology of phylogenetic tree presented in figure 2, the NDV isolates under this study was placed with previously characterized Indonesian and Pakistani strains in genotype VII. Genotype VII is the pre-dominant genotype responsible for recent ND outbreaks in Pakistan. It has been reported before different genotype of NDV (VI and VII) may co-cir- culate in the country and cause disease in domestic and wild birds. Genotype VII is pre-dominantly reported from other Asian countries including China, India, Korea and Taiwan since 1980s.
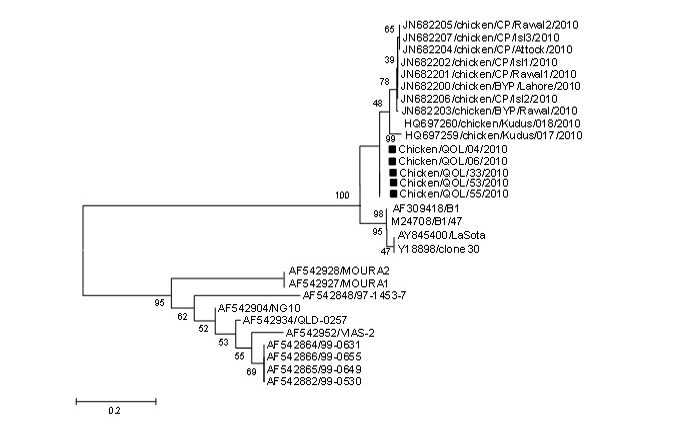
Figure 2: Phylogenetic analysis of HN C-terminal sequences of NDV isolates. The class I and class II viruses are labeled with their predicted HN amino acids extensions. The sequences used in this study are labeled with red color
The study delivers indispensable information on the genetic nature of HN protein of the field NDV isolate and highlights the significance of study on disease diagnosis and control in the region.
REFERENCES




