Journal of Infection and Molecular Biology
Research Article
Sequence Analysis, 3-Dimensional Protein Modelling and Epitope Prediction of Oma87 Gene Orignating from Pasteurella multocida Isolate of Sheep
Kumar Ragavendhar, Palanisamy Prabhakar, Ardhanary Thangavelu, John John Kirubaharan, Navamani Daniel Joy Chandran
Department of Veterinary Microbiology, Madras Veterinary College, Chennai – 600 007, India.
Abstract | Pasteurellosis, caused by Pasteurella multocida, is an acute septicaemic disease characterized by high morbidity and mortality. Apart from cattle and buffaloes, it also results in severe economic losses in sheep and goat. Frequent reports of sheep pasteurellosis in India raised the need for specific and effective vaccination against sheep pasteurellosis. To identify antigenic determinants suitable for epitope based subunit vaccine, we have reported genome and protein sequence based characterization of outer membrane antigen 87 (Oma87) of P. multocida of sheep origin. Oma87 gene was amplified by PCR, purified and sequenced. Sequence analysis showed 99% homology with Pm70 and other P. multocida Oma87 genes available in the GenBank. Later, the 3-dimensional structure, T cell and B cell binding epitopes of the Oma87 protein were predicted using the Oma87 protein sequence. Our studies suggest that predicted T cell binding epitope (FYQYYSAGGIGSLRG) could be used either alone or together with predicted B cell epitope as a suitable epitope based subunit vaccine against P. multocida infection.
Keywords | P. multocida Oma87 gene, Nucleotide sequence, Sequence analysis, Protein modelling, T cell epitope.
Editor | Tahir Yaqub, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | October 14, 2014; Revised | October 30, 2014; Accepted | October 31, 2014; Published | November 15, 2014
*Correspondence | Ardhanary Thangavelu, Madras Veterinary College, Chennai, India; Email: thangavelu_a@hotmail.com
Citation | Ragavendhar K, Prabhakar P, Thangavelu A, Kirubaharan JJ, Chandran NDJ (2015). Sequence analysis, 3-dimensional protein modelling and epitope prediction of Oma87 gene orignating from Pasteurella multocida isolate of sheep. J. Inf. Mol. Biol. 3(1): 1-12.
DOI | http://dx.doi.org/10.14737/journal.jimb/2015/3.1.1.12
ISSN (Online) | 2307-5465; ISSN (Print) | 2307-5716
Copyright © 2015 Ragavendhar et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
The bacterium, Pasteurella multocida (P. multocida), is an opportunistic pathogen responsible for fowl cholera in poultry, haemorrhagic septicaemia of buffalo, pneumonia of lambs and goats, respiratory atrophic rhinitis of swine and purulent rhinitis of rabbit (Gyles et al., 2004). Sheep pasteurellosis, caused by P. multocida serotype A, is one of the common infectious diseases occurring in temperate and sub-tropical areas (Chandrasekaran, 1991). In India, efforts are being done to control this disease with the use of killed vaccine prepared from P. multocida P52 strain. Even though the same vaccine is used for sheep and goats, the outbreaks stills occurs in India as reported previously (Prabhakar et al., 2010; Dar et al., 2012; Prabhakar et al., 2012). This urges the development of newer and more effective vaccines to be used for prophylaxis against pasteurellosis. In this regards, with the development of reverse vaccinology, vaccines could be developed with the aid of computational analysis of genome (Amol et al., 2013).
Since immunization with lipopolysaccharides alone induced only partial protection, outer membrane proteins are thought to be the major antigens involved in inducing cross-protective response (Adler and Adler, 1991). Several outer membrane proteins (OMP) are immunogens and the antibodies against them demonstrate a strong protective activity. Such antigens may be used as a component of a subunit vaccine which lacks the demerits of conventional vaccine (Confer et al., 2001). Further, an 87-kDa outer membrane antigen coded by Oma87 gene has been reported in all 16 serotypes of P. multocida (Raffolo and Adler, 1996). Added to this, Singh et al. (2011) have investigated outer membrane protein DNA for their potency to be used as successful vaccine against haemorrhagic septicaemia of cattle and buffaloes (Singh et al., 2011). In this study, we report the sequencing and sequence analysis of gene encoding 87-kDa Oma87 gene of P. multocida serotype ‘A’ isolate obtained from sheep as well as the generation of 3D protein structure of Oma87 protein with validation by VADAR and Ramachandran plot. We also employed reverse vaccinology based computational analysis for prediction of Oma87 protein specific B cell and T cell epitopes along with their docking studies to the MHC class-I and class-II alleles for the suitable vaccine development.
MATERIALS AND METHOD
P. multocida Isolates
Thirteen P. multocida sheep isolates were inoculated in BHI broth and incubated at 37oC overnight. After incubation all the thirteen isolates were injected (0.2mL) via peritoneal route into thirteen Swiss albino mice and the mean death time was observed. Aspirated heart blood from dead mice was streaked on 10% sheep blood agar and incubated at 37oC. Biochemical tests were carried out for identification. The biochemical tests included IMVIC, sugar fermentation test and catalase and oxidase test (OIE manual, 2008).
DNA Extraction and P. multocida Species Specific PCR (PM-PCR)
DNA was extracted from the culture by high salt method as described previously (Kumar and Ramdass, 2001) and stored at –20oC until used. Previously developed P. multocida specific PCR assay (Townsend et al., 1998) was used for the further confirmation of 13 P. multocida isolates.
PCR and Sequencing of Oma87 Gene of P. multocida
From the thirteen isolates, one P. multocida sheep isolate was selected for amplification of Oma87 gene. Both forward 5´actcacaggataacattcacg 3´ and Reverse 5´ ttttacataagttccaatcgc 3´ primers were designed based on the Oma87 gene sequence (GenBank Accession No. U60439). The PCR reaction mixture contained 50ng of gDNA, 10 pmol of each primer and 1 units of hot start Taq polymerase. The thermal cycle protocol were as follows: Initial denaturation at 94oC for 5 min, followed by 30 cycles, each cycle consisting of 3 steps- denaturation at 94oC for 1 min, annealing at 49oC for 1 min, Extension at 72oC for 3 min. Final Extension was carried out at 72oC for 10 min. The amplified (2.42kb) product was purified and the sequencing was carried out by primer walking. Sequence was submitted to NCBI GenBank (Accession number KJ576825).
Sequence Analysis and Protein Modelling of Oma87 Protein
A global blast analysis of the selected sequence revealed the closest match of it. About nine hits and representative sequences were selected from Blast results for the alignment. The sequences were aligned by clustal w and phylogenetic tree was constructed based on the clustal w alignment using MEGA 6 (Tamura et al., 2004; Tamura et al., 2013). This Oma87 protein sequence was subjected to protein structure modelling using SWISS–MODEL homology modeller (http://beta.swissmodel.expasy.org/workspace) in an automated mode (Benkert et al., 2011). Validation was done by VADAR (Willard et al., 2003) and Ramachandran plot using PROCHECK (Ramachandran et al., 1963; Laskowski et al., 1993). Physiochemical characterization was done by PROTPARAM software using ExPASyserver (Ikai et al., 1980; Kyte and Doolittle, 1982; Guruprasad et al., 1990; Gasteiger et al., 2005). The server VaxiJen (Doytchinova et al., 2007) was used to analyse the antigenicity of the Oma87 protein.
T Cell and B Cell Epitope Prediction, 3D Structure Prediction of T Cell Epitopes, MHC Class II H2-IAB Allele, MHC Class I H2-Kda Allele and their Docking Studies Oma87 protein sequence was subjected to Mouse MHC class I allele and MHC class II allele binding T cell epitope prediction using T cell epitope prediction by IEDB-AR tools (tools.immuneepitope.org/mhc). The epitope having IC50 values within 250nm were selected and their antigen score was analysed using VaxiJen server. Oma87 protein sequence was subjected to B cell epitope prediction using ABC prediction server (Saha and Raghava, 2006). 3D structure of the selected epitope was predicted using PEP-fold 1.5 peptide structure prediction server at the RPBS Mobyle Portal (Maupetit et al., 2009; Maupetit et al., 2010; Thevenet et al., 2012). Best conformation given by the server was selected for further docking studies. Also the 3D structure of Mouse MHC class I H2-kDa allele and Mouse MHC class II H2-I-Ab allele were predicted using SWISS–MODEL homology modeller. Validation was done by Vadar and Ramachandran plot. For this purpose, protein sequence of both H2-kDa allele and H2-I-ab allele were obtained from NCBI gene bank database AAX98169.1 and NP_996988.2 respectively. The docking of the predicted Oma87 T cell epitopes to MHC I H2-kDa Protein and MHC II H2-I-ab protein were carried out using Patchdock server (Schneidman-Duhovny et al., 2005) with the RMSD value 1.5A as recommended for protein small molecule docking and the solved structures were further refined using Firedock server (Mashiach et al., 2008).
RESULTS AND DISCUSSION
Pasteurellosis is an acute septicaemic disease characterized by high morbidity and mortality in cattle, sheep, goat and poultry resulting in economic losses. Pasteurellosis in sheep is capsular A serotype specific, which affects all age group of sheep leading to unthriftiness, reduction in body weight and death (Prabhakar et al., 2010). To develop an effective epitope based vaccine for sheep pasteurellosis, we focused on outer membrane protein of P. multocida which is proved to be immunogenic. Recently, a study based on the immunoinformatics revealed 129 immunogens in P. multocida. Further in vitro gene expres sion studies resulted in twelve immunogenic determinants including Oma87, Omp A, Omp16 and Plp B against P. multocida infected chicken sera (Al-Hasani et al., 2007). Recombinant Oma87 protein developed against Fowl cholera causing P. multocida A serotype showed effective protection in an experimental study with mice and rabbit (Raffolo and Adler, 1996). Immunization of mice with recombinant Oma87 protein developed against P. multocida P52 strain B serotype showed increased antigen specific IgG titres in serum, whereas challenge studies showed 66.6 and 83.3% protection against P. multocida B: 2 strain and P. multocida A: 1 strain, respectively (Kumar et al., 2013).
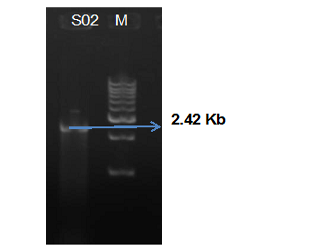
Figure 1: Agarose gel electrophoresis analysis showing Oma87 gene (2.42 Kb) of P. multocida sheep S02 isolate and 1Kb ladder
Table 1: History of the thirteen isolates of P. multocida with origin, date of sample collection and their respective clinical symptoms
|
Isolates |
MVC S01-S04 |
MVC S05-S07 |
MVC S08-S10 |
MVC S11-S13 |
|
Location of outbreak/ sample collection |
Dharmapuri, Tamil Nadu |
Dindigul, Tamil Nadu |
Kattupakkam, Tamil Nadu |
Sunampet, Tamil Nadu |
|
Date of sample collection |
24-06-08 |
25-06-08 |
12-07-08 |
24-12-08 |
|
Species affected |
Ovine |
Ovine |
Ovine |
Ovine |
|
Age of animals |
1-3 years |
1-3 years |
1-3 years |
1-3 years |
|
Clinical symptoms |
Respiratory distress and nasal discharge |
Nasal discharge |
Respiratory distress and nasal discharge |
Nasal discharge |
|
Mouse bio assay (MDT) |
S01-15 hour S02- 13 hour S03- 15 hour S04-17 hour |
S05-15 hour S06- 17 hour S07- 16 hour |
S08-15 hour S09- 16 hour S10- 16 hour |
S11-20 hour S12- 21 hour S13- 28 hour |
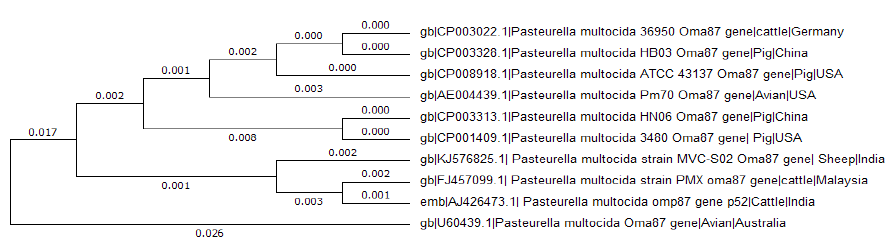
Figure 2: Phylogenetic tree analysis for the Oma87 gene sequence of P. multocida sheep S02 isolate with nine similar sequence obtained by BLAST hits using MEGA6
Based on the conventional microbiology techniques all the 13 isolates were identified as P. multocida (OIE manual, 2008) and further confirmed by P. multocida species specific PCR (PM–PCR) assay (Townsends et al, 1998). In mouse biological assay ten P. multocida isolates were found to be virulent with MDT between 12-18 hrs. Two isolates showed MDT between 19 and 24 hrs. One isolate showed MDT between 25 and 42 hrs (Table 1).
Amplification of Oma87 gene from P. multocida (SO2) isolate resulted in the amplified product of 2.42bp (Figure 1). The amplified product was sequenced by primer walking. Only the complete 2373bp sequence of oma87 gene was submitted to NCBI GenBank under the accession number KJ576825 from the obtained product. It showed 99% homology with P. multocida subsp P. multocida str. Pm70, P. multocida omp87 gene for omp87 protein, strain P52, P. multocida subsP. multocida str. HN06, and complete genome. Clustal w multiple sequence alignment was performed for nine sequences showing best hit with sheep isolate Oma87 gene sequence and phylogenetic tree was constructed using MEGA 6 (Figure 2). Oma87 gene from P. multocida (S02) isolate was found group with P. multocida P52 P. multocid and PMX strains of cattle origin indicating close phylogenetic relationship.
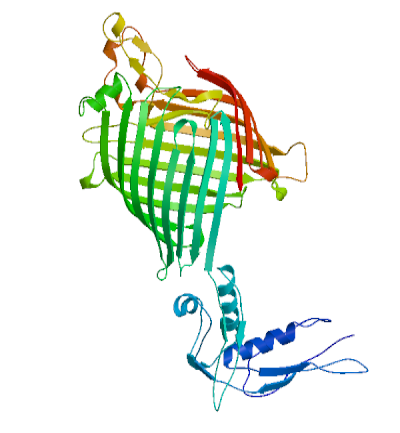
Figure 3: 3D structure of Oma87 protein of P. multocida sheep isolate predicted using Swizz modeller
Table 2: VADAR validation DATA of the predicted 3D model of the Oma87 protein
|
Statistic |
Observed |
Expected |
|
Helix |
107(13%) |
- |
|
Beta |
368(47%) |
- |
|
Coil |
297(38%) |
- |
|
Turn |
96(12%) |
- |
|
Hydrogen bonds |
||
|
Mean HBond distance |
2.3 |
2.2 |
|
Mean HBond Energy |
-1.6 |
-2.0 |
|
Re with Hbonds |
525(68%) |
579(75%) |
|
Volume |
||
|
Total volume(packing) |
105946.0 |
104483.6 |
|
Mean residue volume Mean frac volume |
137.2 1.1 |
125.0 1.0 |
|
Molecular weight |
85680.2 |
- |
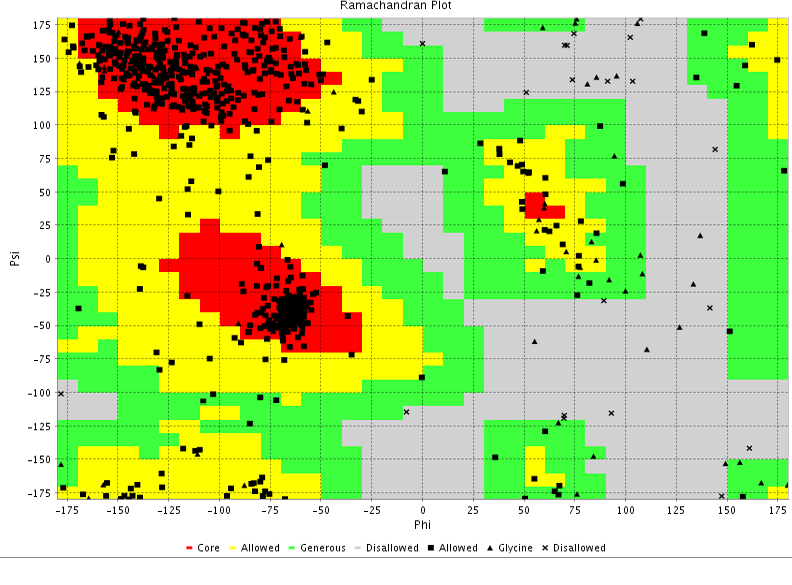
Oma87 protein sequence was used for protein modelling using SWISS–MODEL homology modelling. From the SWIZZ-MODEL templates library, the template that matched Oma87 protein sequence was searched with BLAST and HHBlits and overall 82 templates were found. Oma87 protein sequence showed 59.92% identity with outer membrane protein assembly factor BamA structure (protein data bank code 4k3c, chain A) from Haemophilus ducreyi and the same was used as template for model building.3D Model for Oma87 protein (Figure 3) was built based on the target-template alignment using ProMod and then the model quality was assessed using the QMEAN scoring function of -7.64. The Oma87 protein structure was validated using VADAR (Table 2) and RAMACHANDRAN plot (Figure 4). VADAR validation data showed that the predicted protein contained 13% helix, 47% beta, 38% coil, 12% turn and molecular weight of 85.7 kDa. Ramachandran plot statistics obtained from the PROCHECK analysis revealed that 77.4% of amino acids fall in most favoured region, 16% in additionally allowed region, 3.9 % in generously allowed region and 2.7 % in disallowed region. The physiochemical parameters of the Oma87 protein were computed using the Expasy’s ProtParam tool. The computed pI value was 5.94 (pI<7) indicates the acidic nature of the Oma87 protein. Instability index value is computed to be 30.11 and this classifies the Oma87 protein as a stable protein. The aliphatic index value is 79.97 shows that Oma87 protein may be stable at wide range of temperature. Grand average of hydropathicity index (GRAVY) indicates the solubility of the proteins, for Oma87 protein it was found to be -0.387 showing that the protein was hydrophilic in nature. VaxiJen classified Oma87 protein as probable antigen because it has a value of 0.5514 which is above the normal threshold value of 0.4, proved that Oma87 protein can act as the immunogenic protein.
Table 3: Predicted T cell epitopes of Oma87 protein binding with MHC class I alleles and their antigen score
|
Peptide |
Position |
MHC class I Allele |
IC50 |
Vaxijen score |
|
VGYTYNKL |
524-531 |
H-2-Kb |
2 |
0.5413 |
|
DEQDKTISLDFI |
325-336 |
H-2-Kk |
14 |
0.8986 |
|
SYQASIKQDNFL |
433-444 |
H-2-Kd |
16 |
0.6612 |
|
GYFPTKGVRANI |
574-585 |
H-2-Kd |
23 |
1.0916 |
|
VEAGKSYTVRQI |
337-348 |
H-2-Kk |
23 |
0.8904 |
|
AAYGRTSYGGNL |
497-508 |
H-2-Kb |
27 |
0.8941 |
|
RYNAKVEAI |
143-151 |
H-2-Kd |
31 |
1.03 |
|
DEIEQFQFSI |
777-786 |
H-2-Kk |
34 |
1.5781 |
|
IGYGTESGL |
424-432 |
H-2-Kb |
36 |
1.4592 |
|
SYCLVSSDV |
672-680 |
H-2-Kd |
43 |
0.9801 |
|
RYNAKVEAIVNTL |
143-155 |
H-2-Kd |
44 |
0.4937 |
|
VALQWQSPI |
752-760 |
H-2-Kb |
58 |
0.5112 |
|
AAYGRTSYGGNLTL |
497-510 |
H-2-Kb |
59 |
0.9513 |
|
QERNTGSINFGI |
413-424 |
H-2-Kk |
70 |
1.1842 |
|
KYQGDEIEQFQFSI |
773-786 |
H-2-Kd |
79 |
1.0124 |
|
INFGIGYGTESGL |
420-432 |
H-2-Kb |
88 |
1.1467 |
|
MELQTDAWWKLF |
194-205 |
H-2-Kk |
96 |
0.9976 |
|
FDEQDKTISLDFI |
324-336 |
H-2-Kk |
102 |
1.0353 |
|
KEGDLYTVKCARI |
256-268 |
H-2-Kk |
107 |
1.7008 |
|
QYYSAGGIGSL |
640-650 |
H-2-Kd |
121 |
0.4361 |
|
GYAQFQILDTDI |
229-240 |
H-2-Kd |
149 |
0.6808 |
|
SYQASIKQDNFLGM |
433-446 |
H-2-Kd |
160 |
0.4972 |
|
FYQYYSAGGIGSL* |
638-650 |
H-2-Kd |
198 |
0.4882 |
|
LSYQASIKQDNFL |
432-444 |
H-2-Kb |
200 |
0.7496 |
|
QYDDVKASREGNTL |
68-81 |
H-2-Kd |
201 |
1.3941 |
|
IPDEAIKQNL |
102-111 |
H-2-Ld |
207 |
0.8153 |
|
ISASFADGFGGKRL |
623-636 |
H-2-Kb |
210 |
1.0221 |
|
IPGSDNKYYKL |
591-601 |
H-2-Ld |
221 |
0.6592 |
|
AYGRTSYGGNLTL |
498-510 |
H-2-Kd |
242 |
0.9579 |
* Epitope selected for docking studies
Table 4: Predicted T cell epitopes of Oma87 protein binding with MHC class II alleles and their antigen score
|
Peptide |
Position |
MHC class II Allele |
IC-50 |
Vaxijen Score |
|
FGSTTAFAAPFVVKD |
11-25 |
H2-Iab |
40.6 |
0.5836 |
|
GSLRGFAYGAIGPNA |
648-662 |
H2-IAb |
59.7 |
0.3174 |
|
LPFYQYYSAGGIGSL |
636-650 |
H2-IAb |
75.7 |
0.5686 |
|
PFYQYYSAGGIGSLR |
637-651 |
H2-IAb |
76.4 |
0.6307 |
|
FYQYYSAGGIGSLRG* |
638-652 |
H2-IAb |
84.1 |
0.6434 |
|
LGERGYATAQVNVHP |
308-322 |
H2-IAb |
94.8 |
1.0491 |
|
GERGYATAQVNVHPT |
309-323 |
H2-IAb |
94.9 |
0.8047 |
|
STTAFAAPFVVKDIR |
13-27 |
H2-IAb |
97.7 |
0.4277 |
|
QIRFEGNTSSADSTL |
347-361 |
H2-IAb |
101 |
1.4531 |
|
RQIRFEGNTSSADST |
346-360 |
H2-IAb |
105.1 |
1.6825 |
|
ERGYATAQVNVHPTF |
310-324 |
H2-IAb |
105.1 |
0.8262 |
|
KLGERGYATAQVNVH |
307-321 |
H2-IAb |
114.1 |
0.8048 |
|
VRQIRFEGNTSSADS |
345-359 |
H2-IAb |
120.2 |
1.3336 |
|
VTASTELIVPTPFVA |
687-701 |
H2-IAb |
124.9 |
0.7166 |
|
TTAFAAPFVVKDIRV |
14-28 |
H2-IAb |
127.5 |
0.7409 |
|
IRFEGNTSSADSTLR |
348-362 |
H2-IAb |
128.4 |
1.5361 |
|
RGYATAQVNVHPTFD |
311-325 |
H2-IAb |
149.8 |
0.8189 |
|
TVRQIRFEGNTSSAD |
344-358 |
H2-IAb |
152.2 |
1.2743 |
|
FEEYDSSKSNTSAAY |
485-499 |
H2-IAb |
153.8 |
1.2213 |
|
FFEEYDSSKSNTSAA |
484-498 |
H2-IAb |
158.6 |
1.117 |
|
VFFEEYDSSKSNTSA |
483-497 |
H2-IAb |
186.9 |
1.0261 |
|
ELIVPTPFVADKNQN |
692-706 |
H2-IAb |
188.3 |
0.4855 |
|
RLPFYQYYSAGGIGS |
635-649 |
H2-IAb |
195.1 |
1.0154 |
|
RFEGNTSSADSTLRQ |
349-363 |
H2-IAb |
226.3 |
1.4227 |
|
NVFFEEYDSSKSNTS |
482-496 |
H2-IAb |
238.2 |
0.9596 |
|
EEYDSSKSNTSAAYG |
486-500 |
H2-IAb |
256.3 |
1.271 |
* Epitope selected for docking studies

Figure 6a: 3D structure of MHC class I H2-kDa protein
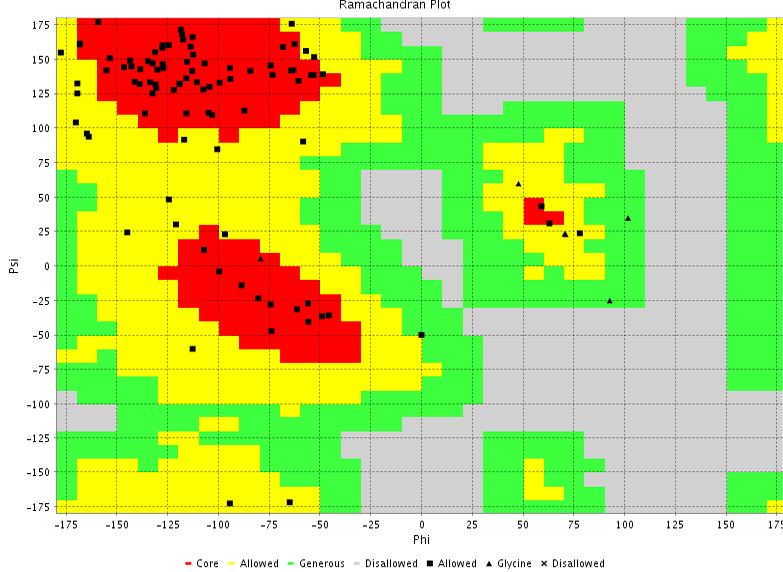
Figure 6b: Ramachandran plot
In order to increase the immune response against the pathogen, activation of Helper T cells and cytotoxic T cells is necessary. Epitope is an antigenic determinant that plays an important role in immunity of an organism (Ansari and Raghava, 2010). Epitope that binds with MHC class I and class II alleles are presented to Helper T cells and cytotoxic T cells for their activation. Hence, the prediction of T cell epitopes that binds with both MHC class I and MHC class II alleles will lead to the development of effective vaccine against P. multocida infection. Since prediction of sheep MHC class I and class II allele binding T cell epitope is not possible with the presently availableprediction server, Mouse MHC class I and class II binders were used for this study. Previous study reported that mouse MHC binding T cell epitopes predicted from the Blue tongue virus non-structural protein NS1 showed increase in IFN-γ production and CTL activity in mice, also proved to be consistently immunogenic in sheep infected with blue tongue virus using IFN-γ elispot assay (Rojas et al, 2014). Prediction of T cell epitope binding with mouse MHC class I alleles (H2KB, H2Dd, H2Kb, H2Kd, H2Kk, H2Ld) and MHC class II alleles (H2IAb, H2IAd, H2IEd) were done by the prediction server. A total of 56 epitopes binding with MHC class I alleles having IC50 value < 250nm was obtained. Likewise, 59 epitopes of MHC class II binders with IC50 value < 250nm was also obtained. Epitopes having binding affinity threshold below 250nm were selected based on the previous study (Sakib et al., 2014). In order to select the best T cell epitope their antigenicity was cross-checked using VaxiJen server, epitope having antigen score above the threshold value of 0.4 were selected, nearly 29 epitopes binding with MHC class I alleles and 26 epitopes binding with MHC class II alleles satisfied both the criteria of IC50 value <250nm and Antigen score >0.4 (Table 3 and Table 4). Further analysis showed that the MHC class I binding epitope KEGDLYTVKCARI had the highest antigen score of 1.7 and RQIRFEGNTSSADST epitope had the highest antigen score of 1.6 in the case of MHC class II binders. Best T cell epitope should be able to bind with both MHC class I and MHC class II alleles, out of 29 epitopes from MHC class I H2-kDa binding FYQYYSAGGIGSL and QYYSAGGIGSL epitopes share their sequence with MHC class IIH2-IAb binding FYQYYSAGGIGSLRG and PFYQYYSAGGIGSLR epitopes. From these FYQYYSAGGIGSLRG epitope had higher antigen score of 0.6434 showed sequence similarity with both MHC class I binding FYQYYSAGGIGSL and QYYSAGGIGSL epitopes, whereas FYQYYSAG
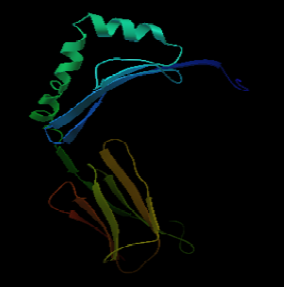
Figure 7a: 3D structure of MHC class II H2-IAb protein
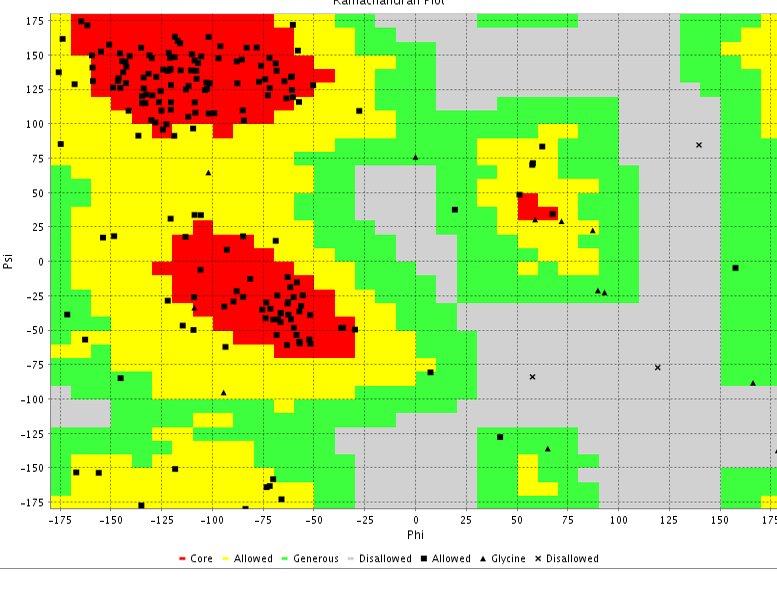
Figure 7b: Ramachandran plot
GIGSL epitope showed higher antigenic score of 0.4882 compared to the score of QYYSAGGIGSL epitopes (0.432). Based on this FYQYYSAGGIGSLRG and FYQYYSAGGIGSL were selected for further docking studies with MHC class II H2-IAb allele and MHC class I H2-kDa allele. Sequence similarity search of these epitopes was done using BLAST against Mus musculus, Ovis aries and Gallus gallus; no similarity was found which showed the uniqueness of the epitopes. B cell epitopes including linear and conformational structure mostly induce humoral immune response (Conway et al., 2003; Malm et al., 2005). Twenty B cell epitopes of Oma87 protein was predicted using ABC prediction server (Table 5). Of these EGNTSSADSTLRQEMRQQEG epitope shows high antigenic score of 1.5 and epitope ADGFGGKRLPFYQYYSAGGI with antigen score of 0.8 shares partially similar sequence with FYQYYSAGGIGSLRG and FYQYYSAGGIGSL epitopes.
Table 5: Predicted B cell epitopes of Oma87 protein and their respective antigen score
|
Position |
Epitope |
Score |
|
81 |
LVVTVMPKPVISNVVIDGNK |
0.34 |
|
488 |
YDSSKSNTSAAYGRTSYGGN |
1.34 |
|
351 |
EGNTSSADSTLRQEMRQQEG |
1.59 |
|
149 |
EAIVNTLPNNSAEIKIQINE |
0.897 |
|
581 |
VRANIGGRVTIPGSDNKYYK |
0.465 |
|
26 |
IRVDGVQAGTEGSVLATLPV |
1.33 |
|
384 |
RLDRTGFFESVETKTEAIPG |
0.04 |
|
761 |
GPLVSSYAKPLKKYQGDEIE |
0.164 |
|
724 |
WKAEDKAKFAKLNVPDYSDP |
0.29 |
|
243 |
SDDKKEARVIIKVKEGDLYT |
0.79 |
|
313 |
YATAQVNVHPTFDEQDKTIS |
0.68 |
|
412 |
SQERNTGSINFGIGYGTESG |
1.28 |
|
513 |
PVNENNSYYLGVGYTYNKLK |
0.35 |
|
535 |
APEYNRDLYRQSMKYNDSWT |
0.46 |
|
602 |
NAEAQGFYPLDREHGWVLSS |
0.09 |
|
680 |
VIGGNAMVTASTELIVPTPF |
0.75 |
|
191 |
EDQMELQTDAWWKLFGNKFD |
0.92 |
|
628 |
ADGFGGKRLPFYQYYSAGGI |
0.80 |
|
445 |
GMGSSISLGGTRNDYGTTIN |
1.26 |
|
659 |
GPNAIYRTRQCPDSYCLVSS |
0.08 |
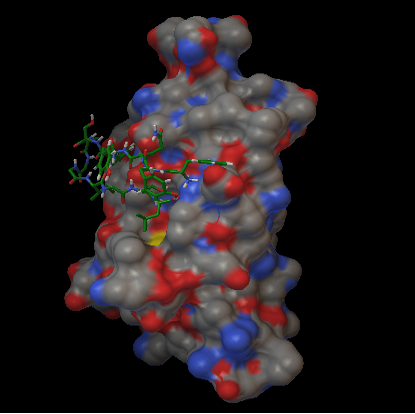
The 3D structure of the FYQYYSAGGIGSLRG and FYQYYSAGGIGSL epitopes was predicted using PEP fold structure prediction server. Best model proposed by the server was selected for docking studies (Figure 5a and 5b). 3D structure of MHC class I H2-KDa allele and MHC class II H2-IAb allele were predicted by Swiss-modeller using the protein sequence obtained from NCBI database AAX98169.1 and NP_996988.2 respectively.3D model of MHC class I H2-KDa (Figure 6) was built using the template H-2 class I histocompatibility antigen, D-37 alpha chain (3vj6.1). Sequence identity of the template to the MHC class I H2-kDa was 90% and the quality was accessed by Qmean scoring function of -0.83.The template 3K peptide, linker and MHC H-2 class II I-Ab beta chain (3rdt.1.B) was used to build the3D model of MHC class II H2-IAb (Figure 7). Sequence identity of the template to the query sequence was 98% and Q mean score was -0.92.3D model of both MHC class II H2-IAb allele and MHC class I H2- kDa allele was validated using VADAR (Table 6) and RAMACHANDRAN plot (Figure 6 and 7).
Table 6: VADAR validation of 3D structure of MHC class I H2-kDa protein and MHC class II H2-IAb protein
|
Statistics |
MHC Class I-H2-Kd |
MHC Class II-H2-IAb |
|
Helix |
0 |
28(14%) |
|
Beta |
61(61%) |
91(46%) |
|
Coil |
38(38%) |
76(38%) |
|
Turn |
24(24%) |
28(14%) |
|
Hydrogen bonds |
||
|
Mean HBond distance |
2.2 |
2.2 |
|
Mean HBond Energy |
-1.7 |
-1.7 |
|
Re with Hbonds |
60(60%) |
132(67%) |
|
Volume |
||
|
Total volume(packing) |
13010.3 |
25968.4 |
|
Mean residue volume Mean frac volume |
131.4 1.0 |
133.0 1.0 |
|
Molecular weight |
11388.26 |
22894.34 |
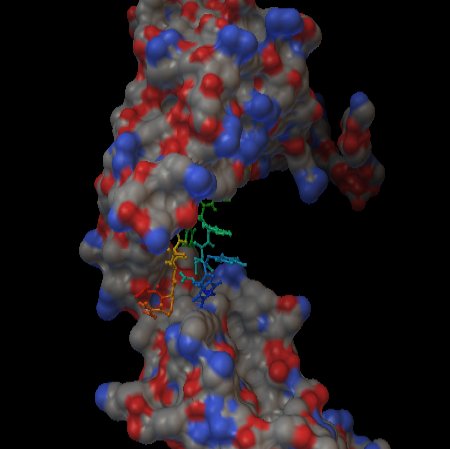
Using PATCHDOCK server binding model of selected T cell epitopes with their respective MHC class I and MHC class II alleles were generated and their refinement was done by FIREDOCK server. FYQYYSAGGIGSLRG epitope was found to bind to the MHC class II H2-IAb allele with the binding score of 5918 and binding energy of -34.09 (Figure 8 and Table 7). In the case of FYQYYSAGGIGSL epitope-MHC class I H2-kDa allele (Figure 9) the binding score and the binding energy were 8238 and -60.15 respectively (Table 7). Based on the binding affinity with MHC class II allele, antigenic score and the docking result FYQYYSAGGIGSLRG epitope which could be superimposed with MHC class I allele binding FYQYYSAGGIGSL epitope may be a suitable vaccine candidate against sheep Pasteurellosis. Longer duration of vaccine- mediated immunity can be achieved by the combination of both humoral and cell mediated immunity (Amanna IJ and Slifka MK, 2011).Hence, we suggest that the B cell epitope ADGFGGKRLPFYQYYSAGGI which had partially similar sequence with predicted T cell epitope can be coupled for inducing antibody response and cell mediated immunity. In this present work, using computational analysis we predicted both T cell and B cell binding epitopes of Oma87 protein with lesser time which may consume more time and extensive trial if carried out by experimental approaches. Further in vivo work with the predicted epitopes in animals would show their ability to induce protective immune responses against P. multocida.
Table 7: Binding affinity of FYQYYSAGGIGSL epitope with MHC class I H2-kDa protein and FYQYYSAGGIGSLRG epitope with MHC class II H2-IAb protein
|
Epitope |
MHC alleles |
Global energy |
Binding score |
Attractive VdW |
Repulsive VdW |
ACE |
HB |
|
FYQYYSAGGIGSL |
Class I H2-kDa |
-26.72 |
5390 |
-26.93 |
12.17 |
-5.04 |
-2.95 |
|
FYQYYSAGGIGSLRG |
Class II H2-I-AB |
-60.15 |
8238 |
-32.03 |
20.38 |
-15.50 |
-1.57 |
ACKNOWLEDGEMENTS
Sincere thanks are due to ICAR, New Delhi and Dean, Madras Veterinary College, Chennai for providing necessary facilities.
REFERENCE