Journal of Infection and Molecular Biology
Research Article
Mitochondrial ATPase 6,8 Associated with Brain Tumours in Patients Compared to Adjacent Normal Brain Cells
Massoud Houshmand1, Soha Sadeghi2, Donya Altafi2, Mahsa Aliqanbari1, Hamed Hojatian2*
1National Institutes for Genetic Engineering and Biotechnology, Tehran, Iran; 2Department of Cellular and Molecular Biology, Nour Danesh Institute of Higher Education, Isfahan, Iran.
Abstract | Background: Brain cancer is considered one of the most prevalent types of cancer in the world. Primary brain tumours consist of two types. Studies provide some deficiencies in mitochondrial functions that could cause different genetic. Objective: This study aimed to determine the association of ATPase 6,8 alterations of brain tumour cells in comparison with the adjacent healthy tissue cells. Methods: A group of patients was examined, and their disease was identified during precise examinations. These persons were sampled for their affected brain tissues, and these were compared with their adjacent healthy cells. Besides, the populations of 300 healthy controls were selected as the control. The DNA of the brain tumour cells was extracted and analysed using sequencing methods. Result: After the sequence analysis, T8473C, G8584A, A8701G, A8730G and A8860G variants were found—all of them had been reported in other diseases. Also, they were observed in patients with brain tumours, as compared with the adjacent normal tissues. Discussion: The A8860G variant was one of the most prevalent polymorphisms between all these alterations in brain cancer. It seems that the ATPase 6 subunit is more prone to brain cancer. The analysis shows that amongst all the five variants determined in this research, the T8473C, G8584A and A8730G variants—with the p value<0.05—were considered to affect brain tumours.
Keywords | ATPase6 8, Brain cancer, Tumor, Mitochondria, Polymorphism, MT-ATP6, MT-ATP8
Editor | Tahir Yaqub, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | June 23, 2018; Accepted | August 04, 2018; Published | August 27, 2018
*Correspondence | Hamed Hojatian, Department of Cellular and Molecular Biology, Nour Danesh Institute of Higher Education, Isfahan, Iran; Email: hamedgenesis@gmail.com
Citation | Houshmand M, Sadeghi S, Altafi D, Aliqanbari M, Hojatian H (2018). Mitochondrial atpase 6,8 associated with brain tumours in patients compared to adjacent normal brain cells. J. Inf. Mol. Biol. 6(2): 45-50.
DOI | http://dx.doi.org/10.17582/journal.jimb/2018/6.2.45.50
ISSN (Online) | 2307-5465; ISSN (Print) | 2307-5716
Copyright © 2018 Houshman et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
According to WHO, a primary brain tumour includes 160 types. The most common of these categories are gliomas and medulloblastoma. Gliomas are known by histological standards as consisting of the presence of mitoses, cytoplasmic morphology or devious nuclear, glomeruloid angiogenesis and necrosis. Based on the WHO classification system, gliomas can be categorized into four different grades with increasing malignancy apportion (Rich and Eyler, 2008). Grade 4 (malignant glioblastoma multiform—GBM) is considered to have the highest invasiveness amongst all the grades. GBM can cause different symptoms—for instance, necrosis, excessive vascular proliferation, and rising expression angiogenic factors (Brat et al., 2015, Friedman et al., 2009). The annual prevalence of malignant glioblastoma is about 5 cases per 100,000 people in the world. Also, more than 18,000 new cases are known in the United States each year (Liu et al., 2010, Wen and Kesari, 2008). Treatments of malignant glioblastoma include surgical resection and radiation therapy, with some new studies in chemotherapy drugs, such as Temozolomide, Bevacizumab, Irinotecan, etc. Based on recent studies, simultaneous use of these drugs with other treatments is more effective when compared with radiation therapy alone (Stupp et al., 2005, Vredenburgh et al., 2007). There are many studies about the relevance of the connection between mtDNA and well-known diseases like cancers and diabetes. So, mitochondrial genes could participate in cancers—for example, breast, colon and kidney cancers (Raule et al., 2007).
The human mitochondrion is defined by circular double-strand DNA and double-membrane with independent genome, transcription and translation machinery. Some genes are encoded by mitochondrial DNA, such as 13 genes of oxidative phosphorylation subunits, 22tRNA and 2rRNA genes, and all the mitochondrial assembly factors encoded by nuclear DNA. The mitochondria play an indispensable role in cells, including control of apoptosis, autophagy, reactive oxygen species (ROS), heat production, etc. The most well-known role of the mitochondrion is to supply ATP (adenosine triphosphate) during oxidative phosphorylation system. It demonstrates that mitochondrial DNA mutations could relate to many neurodegenerative diseases and cancers. Primary studies by Otto Warburg suggested the mutation in mitochondrial DNA could affect carcinogenesis processing and tumours. He explained different mechanisms for taking glucose to produce ATP between healthy and cancer cells. The oxidative phosphorylation system (OXPHOS) is essential to some organs—for example, the heart, muscles, and brain—which require high energy. Therefore, it is not surprising that any failure in mitochondrial OXPHOS would cause pathogenic disorders in these organs (Raule et al., 2007, Carew and Huang, 2002, Hejzlarova et al., 2014, Jonckheere et al., 2012, Wong, 2013, Kroemer, 2006, Greaves et al., 2009). Complex V or ATP synthase is assigned to make adenosine triphosphate (ATP). The variants of ATPase subunit 6 and subunit 8 genes have demonstrated in many cancers that the two of these genes pertain to Complex V (Ghaffarpour et al., 2014, Parrella et al., 2001, Chatterjee et al., 2006, Cuezva et al., 2002). According to previous studies, there has been no experience of mitochondrial ATPase 6 and 8 subunits and their correspondence to brain cancers up to now. Thus, these studies are required to investigate the correlation of mitochondrial genes alterations in brain tumours, which improve our understanding of advances in brain cancers. Like the previous studies, we also intend to investigate ATPase subunit 6 and subunit 8 genes to detect their association with brain tumours in comparison with the normal adjacent tissues. In this study, we suggested some ATPase 6,8 alterations that could be associated with brain tumours.
Materials and methods
Samples
The number of patients with brain cancer was gathered from Cancer Institute Center of Lorestan, Iran, and 300 peripheral blood samples from healthy individuals as standard controls from Houshmand Genetic Laboratory of Tehran, Iran. Information related to these patients is given in the table below (Table 1), as can be seen, 92 percent males and 8 percent females with 40% glioblastoma multiform, 36% astrocytomas, 12% oligodendrogliomas, 8% ependymoma and 4% giant cell glioblastoma were included. All tumour tissues were frozen in liquid nitrogen immediately after removal and then stored at -800 degrees C until DNA extraction.
Table 1: Information about the patients with brain cancer, with details given in the table below
| Characteristic | Freq |
|
Male female |
92% 8% |
|
Age < 40 years > 40 years |
72% 28% |
|
Clinic data Right fronto-temporal Left fronto-temporal Right temporal Left temporal Left frontal Right frontal Right parieto-occipital Right fronto-parietal Intraventicular Cerebral |
12% 4% 24% 8% 12% 8% 8% 12% 4% 8% |
|
Diagnostic Astrocytoma Giant Cell Glioblastoma Oligodendroglioma Ependymoma Glioblastoma multiform |
36% 4% 12% 8% 40% |
DNA Extraction and Primers
Genomic DNA was extracted from tumour tissues with the DNeasy 96 Blood & Tissue Kit (21586 and 21581 Nos. CatQiagen GmbH, Hilden, Germany). The forward and reverse primers used in this research were following the previous study (Ahari et al., 2008).
Polymerase Reaction Chain (PCR)
A 1079 bp fragment was amplified for screening alterations into mitochondrial ATPase 6,8 genes. In brief, PCR was performed with 3 µL total DNA, 0.8µL of each primer, 0.5 µL of dNTPs; 2.5 µL of the buffer; 0.8 µL MgCl2; and 0.3 unit Taq polymerase (GenFanavaran company, Tehran, Iran) in the final volume of 25mL. After addition of 10 µL of mineral oil (Sigma) to PCR protocol: primary denaturation, 5 min per-PCR incubation step at 94°C, 35 cycles of denaturation (94°C) for 50 s, annealing (55°C) for 50 s, and extension (72°C) for 50 s and final extension of 72°C for 10 min were carried out using an automated DNA Thermal Cycler (Perkin-Elmer Cetus). The 1079bp fragment was run on 1% agarose gel.
Sequencing Analysis
The PCR products were sequenced on Applied Biosystems
Table 2: All the polymorphisms found in this study are shown in the table below, with all the information about them
| Mutations | P | Ho | HE | Reported | Unreported | Locus | Codon numbers | Nucleotide changes | Curated references |
| 8473 | + | + | - | + | - | MT-ATP8 | 36 | T-C | 16 |
| 8584 | + | + | - | + | - | MT-ATP6 | 20 | G-A | 34 |
| 8701 | + | + | - | + | - | MT-ATP6 | 59 | A-G | 87 |
| 8730 | + | + | - | + | - | MT-ATP6 | 68 | A-G | 5 |
| 8860 | + | + | - | + | - | MT-ATP6 | 112 | A-G |
118 |
Table 3: The table below shows all polymorphisms found in this study, based on the data briefly obtained of p-value, chi-square test, and risk estimates.
| T 8473 C | A8584G | A 8701 G | A8730G | A 8860 G | |
| p value. | 004. | 004. |
469. |
004. | - |
| Chi- Square Tests |
8.333a |
8.333a |
.523a |
8.333a |
- |
| Risk Estimate | 920. | 920. | 704. | 920. | - |
Table 4: Information on normal population in this study
| Variations | p value | Ho | He | Reported | Unreported |
| A 8460 G | .316 | + | _ | + | _ |
| C 8461 T | .316 | + | _ | + | _ |
| T 8462 C | .316 | + | _ | + | _ |
| T 8503 C | .316 | + | _ | + | _ |
|
A 8527 G |
.316 | + | _ | + | _ |
| C 8562 T | .316 | + | _ | + | _ |
| T 8594 C | .316 | + | _ | + | _ |
| T 8614 C | .316 | + | _ | + | _ |
| A 8667 G | .316 | + | _ | + | _ |
| C 8676 T | .316 | + | _ | + | _ |
| A 8679 G | .316 | + | _ | + | _ |
| T 8763 C | .316 | + | _ | + | _ |
| A 8812 G | .316 | + | _ | + | _ |
| A 8828 G | .316 | + | _ | + | _ |
| G 8865 A | .316 | + | _ | + | _ |
| T 8867 C | .316 | + | _ | + | _ |
| G 9053 A | .316 | + | _ | + | _ |
| A 8653 G, | .155 | + | _ | + | _ |
| T 8937 C | .155 | + | _ | + | _ |
| G 8994 A | .155 | + | _ | + | _ |
| G 9055 A | .155 | + | _ | + | _ |
| C 8684 T | .024 | + | _ | + | _ |
| G 8697 A | .043 | + | _ | + | _ |
| A 8701 G* | .469 | _ | + |
+ |
_ |
| A 8836 G | .081 | + | _ | + | _ |
| A 8860 G* | _ | + | _ | + | _ |
ABI 3100 sequencing machine (Thermo Fisher Scientific, Waltham, MA, USA). All the amplicons were sequenced in both directions for the alterations of any mitochondrial ATPase 6,8 variations. It was used of FinchTV software to analyse the variants (PerkinElmer Inc., Waltham, MA, USA).
Statistical Analysis
The statistical analysis was performed by SPSS 21.0 (for Windows) to attain the chi-square and the odds ratio for finding an association between ATPase 6,8 of the brain tumours and the DNAs of blood cells as a control. A P-value of <0.01 was considered statistically significant.
Results
In this study, the complete sequences of ATPase 6 and 8 genes of tumour tissues were analysed, and their association between mitochondrial ATPase 6 and 8 genes and the brain tumours was investigated. The 1079 band lane was observed on agarose gel after analysing the PCR products. The results indicated variations between tumour tissue and normal population in three variants in MT-ATP6 and MT-ATP8 (Figure 1).
Five polymorphisms related to patients with brain tumours were found. These polymorphisms were A8701G, G8584A, T8473C, A8730G, and A8860G. The alterations of these polymorphisms were shown briefly in Table 2.
An analysis of the results showed considerable association with the brain pathogenesis for the variations 8730, 8584 and 8473 (chi-square = 8.333, odds ratio = 0.920, P-value<0.05) (Table 3). These polymorphisms—found in two out of 25 patients—were seen in 8% of the patients. Also, they did not find one in the normal sample. The variation 8701 (chi-square=0.523, odds ratio 0.704, p-value>0.05) did not show a significant association with brain tumour pathogenesis. The variation 8860 was found in 100% of patients and most of the normal population (Figure 2). The results of the normal population in this research are demonstrated in Table 4. All variations in the normal population were homoplasmy except the A8701G variant.
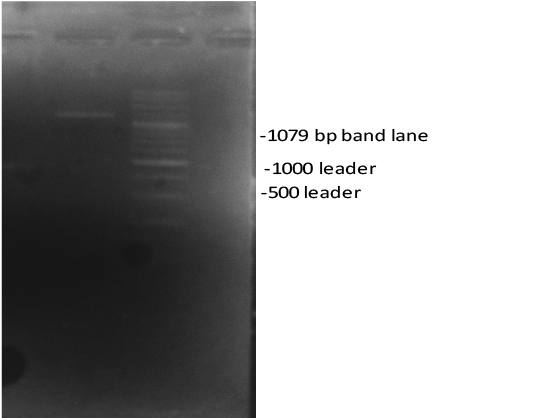
Figure 1: Gel electrophoresis analysis of the mitochondrial ATPase 6 and 8 genes using 1% agarose gel. Notes: lane 1079 indicated the PCR product.
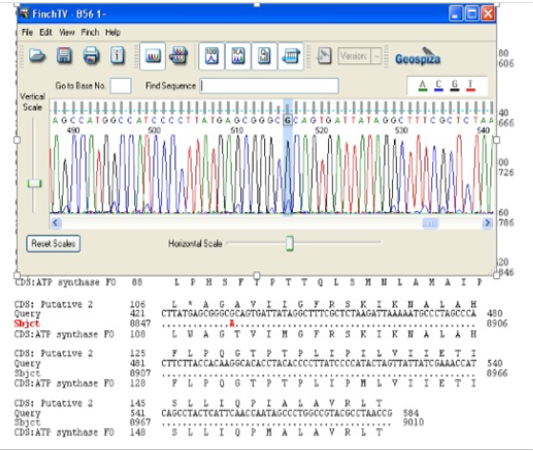
Figure 2: Page of the Finch TV software that demonstrate the A8860G variant, which is marked by red.
Discussion
This study is the first report on the association between ATPase 6,8 genes of mitochondria and the brain tumours. We screened the mitochondrial ATPase 6,8 genes of patients with brain cancer and their susceptibility to ATPase 6,8 alterations. According to Mitomap (http://www.mitomap.org), there are more than 30 mutations and sequence variations in mtDNA associated with brain tumours in mitochondrial genome database that have been reported until now (Yusoff, 2015). The cancer cells, when compared with the healthy human cells, show avarice for glucose uptake from lactate manufacture to supply ATP, which Otto Warburg explained for the first time as maybe existing as an impaired or suppressed function of respiratory enzymes complexes (Lin et al., 2012). Thus, the mitochondria play a significant role in the production of energy for cancer cells in comparison with normal human cells. According to Carew and Huang’s studies, defects in mitochondria can affect many types of cancers and tumours (Carew and Huang, 2002). Based on Shidara et al. and Kirches’s researchers, ATPase 6 gene may increase the probability of cancer progression by aversion cell death pathway (Kirches, 2009, Lu et al., 2009).
ATPase 6,8 genes, the two genes of Complex V enzyme, play an essential role in the respiratory chain of mitochondria (Matsuyama et al., 1998), and have many other crucial roles, in both the normal human cells and cancer cells, such as colonic crypts cancer, breast cancer, thyroid tumour, ovarian cancer, etc. Many studies have indicated the relativity of the mitochondrial ATPase 6,8 and cancers such as ovarian, colon, and kidney and breast cancer (Ghaffarpour et al., 2014).
The results indicated variations between tumour tissue and healthy population in three variants in MT-ATP6 and MT-ATP8 (Figure 1).
Based on the results, the odds ratios are less than 1 and illustrated a low risk of the pathogenesis of these variations. Primary studies for the variations 8473, 8584 and 8730 are explained by Tanaka (1994), Obayashi (1992) and Zhadanov, (2006) respectively. 8701 variant was first reported by Jun et al. (1994), which is probably associated with some diseases like optic neuropathy and dystonia (Jun et al., 1994), Fragmentation of Human Heart Mitochondrial DNA Associated with Premature Ageing (Katsumata et al., 1994), respiratory dysfunction (Lertrit et al., 1994), hypertrophic cardiomyopathy (Obayashi et al., 1992), encephalomyopathies and Parkinson’s (Ozawa et al., 1991). The variant 8860 was observed in colorectal cancer (Taylor et al., 2003, Aikhionbare et al., 2004), ovarian cancer (Houshmand et al., 2011), breast cancer (Matsuyama et al., 1998, Zhu et al., 2005, Sawabe et al., 2011), human glioma cells (Kirches, 2009), osteosarcoma (Aikhionbare et al., 2007), and Leber’s hereditary optic neuropathy (DeHaan et al., 2004). With the P-value of 8860 variant was considered, it probably does not have any significant effect on brain tumour risk. Wei-Feng Wang first explained 8860 variant in 2013, which was also considered as being associated with irritable bowel syndrome with diarrhea (Wang et al., 2013).
8730, 8584 and 8473 are found only inpatient samples, but the 8860 and 8701 variants are found in both patient and healthy populations. The 8730, 8584 and 8473 are probably associated with brain tumour pathogenesis and more susceptive in comparison with other variants that were present only in tumour tissues. Thus, these are not somatic, and we could not say they have significant effects on brain tumour pathogenesis because this used blood for control instead of regular or healthy brain tissue. The 8860 and 8701 variations are not associated with brain tumour pathogenesis when considered with the top data. Our studies suggest that in patients with a brain tumour, ATPase 6 gene probably is more susceptive than ATPase 8 gene.
We require more investigations into other genes of mitochondria regions and also other polymorphisms that are probably associated with brain tumour pathogenesis. The polymorphisms are multiform of DNA sequences, and there are many of them in eukaryotes genome. This multiform is approximately 10000 in number. There are several types of this multiform, such as SNPs, RFLPs, mini-satellite and micro-satellite sequences. Many functions of polymorphism were described, such as gene mapping, assay, and analysis of correlations, fingerprint experiments, and identifications. Thus, there are many reasons to work about polymorphisms, and we hope to investigate and discover many more pieces of astounding information on genes and the world of genetics.
Acknowledgments
We would like to thank all patients and their families for helping us. Thanks also to the National Institute of Genetic Engineering and Biotechnology, Tehran, Iran, and Nour Danesh Institute of Higher Education, Isfahan, Iran, which supported us and cooperated with us to conduct this research.
Conflict of Interest
The authors have no conflict of interest to declare.
Authors Contribution
Hamed Hojatian and Massoud Houshmand conceived of the presented idea and supervised the project. They provided the samples. Hamed Hojatian and Soha Sadeghi reviewed the existing journals’ policy. Donya Altafi and Mahsa Aliqanbari helped oversee the project, and they had participated in study design. Soha Sadeghi, Donya Altafi and Hamed Hojatian performed the statistical analysis. Soha Sadeghi, Donya Altafi and Hamed Hojatian discussed the results and contributed to the writing of the final version of the manuscript. Hamed Hojatian and Donya Altafi carried out all experiments. All authors, participate in revising the article critically for important intellectual content.
References





