Journal of Animal Health and Production
Research Article
Molecular Surveillance of Lumpy Skin Disease Outbreak, 2019 in Sohag, Egypt: Enzootic Potential, Phylogenetic Assessment and Implications on Cattle Herds Health
Hassan Mahmoud Diab1*, Ahmed S. Ahmed2, Gaber El-Saber Batiha3, Luay Alkazmi4 , Mona A. El-Zamkan2
1Department of Animal and Poultry Health and Environment, Faculty of Veterinary Medicine, South Valley University, Qena 83523, Egypt; 2Department of Food Hygiene and Control (Milk Hygiene), Faculty of Veterinary Medicine, South Valley University, Qena 83523, Egypt; 3Department of Pharmacology and Therapeutics, Faculty of Veterinary Medicine, Damanhour University, Damanhour 22511, AlBeheira, Egypt; 4Biology Department, Faculty of Applied Sciences, Umm Al-Qura University, Makkah 21955, Saudi Arabia.
Abstract | Lumpy Skin Disease (LSD) is a remerged threat to livestock health in the Middle East regions that has catastrophic economic implications. LSD has an endemic pattern in Egypt based on the epidemiological surveillances over decades which highlighted the temporal and spatial distribution of the disease episodes. Molecular epidemiology of LSD greatly enhance the genotypic data about the LSD viral strains in Egypt which significantly assets to select the convenient combating strategy to protect cattle herds health. This current study aimed to investigate LSD outbreak occurred in 2019 in Sohag governorate, Egypt among Eight cattle farms of small sized herd reared under byre housing systems. A total of 150 cows belong to 8 dairy cattle herds were examined. Samples including skin biopsy, nasal swabs and saliva dribbling were collected from clinically affected cows and bedding materials from their environments. The samples were used for molecular detection, phylogenetic analysis of LSD virus and identification of the bacterial species incorporated in secondary bacterial infection among infected cattle. Based on descriptive epidemiology of the outbreak, the overall prevalence of lumpy skin virus infection between May to July 2019 among all investigated herds was 25.3%. The intra-herd prevalences were ranged from 16.5% to 37.5%. The most predominating clinical manifestation was the appearance of cutaneous nodules in 75% infected animals. Oedema of fore or hind limbs and other parts of the body including brisket were observed in 26% of cases accompanied with various degree of lameness in 18% of the affected cows. Staphylococcus aureus, E.coli and Klebsiella species were isolated as secondary invaders from all type of samples. Molecular identification of the LSD virus revealed that P32 gene was successfully amplified in 63% of all examined samples. Overall, most of isolates exhibited strong degree of phylogenetic relatedness indicating that strains of LSD virus circulating in Egypt, have well established enzootic state. Further comprehensive analysis of the genetic phylogeny relatedness of LSD virus from Egypt and neighboring countries seem to be crucial to draw conclusive evidences regarding the transboundary threats of LSD to livestock health in Egypt. These will greatly assist to trace back the transmission episodes and to understand the molecular epidemiology of the LSD allover Egypt.
Keywords | Lumpy skin Diseases, Epidemiology, Herd Health, Cattle, Phylogeny
Received | May 17, 2021; Accepted | May 21, 2021; Published | October 01, 2021
*Correspondence | Hassan Mahmoud Diab, Department of Animal and Poultry Health and Environment, Faculty of Veterinary Medicine, South Valley University, Qena 83523, Egypt; Email: hassan.abdelatef@vet.svu.edu.eg
Citation | Diab HM, Ahmed AS, Batiha GS, Alkazmi L, El-Zamkan MA (2021). Molecular surveillance of lumpy skin disease outbreak, 2019 in sohag, egypt: enzootic potential, phylogenetic assessment and implications on cattle herds health. J. Anim. Health Prod. 9(4): 406-416.
DOI | http://dx.doi.org/10.17582/journal.jahp/2021/9.4.406.416
ISSN | 2308-2801
Copyright © 2021 Diab et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Trans-boundary animal diseases including Lumpy skin disease (LSD) are of global concern due to their financial burden on livestock health and industry (Kumar et al., 2021). Hence, the World Organization for Animal Health (OIE) has designated LSD as a notifiable disease (Badhy et al., 2021). LSD was initially discovered in Zambia in 1929 then disseminated to southern and eastern Africa. It was subsequently evolved to Southern African countries, but in the 1970s it spread northwest via the continent into Sub-Saharan West Africa. As of 2000, it has transmitted into many other Middle Eastern countries, and in 2013, it was spread to many more countries around the world (Gibbs 2021). LSD affects cattle of all ages and breeds and has a high morbidity and low fatality rates. Reduced milk yield, meat production, abortion, infertility, condition loss, and hide damage all contribute to severe economic concerns. In the Middle East and Africa, LSD poses threats to bovine health sector (Leliso et al., 2021). The causative agent of LSD is known as lumpy skin disease virus (LSDV) which belongs to genus Capripoxvirus, family Poxviridae. Genome of LSDV is double-stranded DNA of 151 kbp in size that has 156 putative genes. The Capripoxvirus genus includes others important viruses affecting different animal species like sheep pox virus (SPPV) and goatpox virus (GTPV). Those groups of viruses have high degree of homology and found to be phylogenetically related (Tulman et al., 2001). Major route for transmission and spread of LSDV is thought to be via biological vectors especially during summer and humid seasons in tropical and subtropical area. Mosquitoes, mainly like Aedes aegypti, hard ticks like Ixodid species and Culicoides were reported to be incorporated in transmission of LSDV globally (Hussein et al., 2017; Zeedan et al., 2019).
Enzootic occurrences of LSD in Egypt lead to list it among viral infectious endemic diseases that pose alarming concern for cattle health and production (Ahmed et al., 2021). First epizootic attack of LSD was reported in 1988 among cattle population in Eastern regions at Suez and Ismailia governorates, Egypt where two outbreaks had been notified (House et al., 1990). The temporal patterns of LSD were reported to be more frequent and intense during summer every year with wide spatial dissemination over districts of Egypt in each episode. This endemic state was reported even in the presence of annual mass vaccination for susceptible host using vaccine derived from sheep pox strain (Allam et al., 2020). In early 2006, LSD outbreak has invaded cattle in different localities of Egypt, exerting severe economic losses to livestock industry (El-Kholy et al., 2008). Since that, Multiple outbreaks have reemerged in different Egyptian provinces between 2006 to 2019 (Abd El-Hady et al., 2006; El-Kholy et al., 2008; Amal et al., 2010; Salib and Osman, 2011; El-Nahas et al., 2011; Amin et al., 2015; Lamya et al., 2017; Nashwa et al., 2017; Abdallah et al., 2018; Zeedan et al., 2019; Ahmed et al., 2021). The disease was associated with high prevalence of insect vectors which facilitate the transmission of LSDV. Animals are mostly exposed to insect vectors due to defects in the housing system and animal husbandry in cattle farms (Sharawi and Abd El-Rahim, 2011).
LSD is an infectious, eruptive, occasionally fatal disease of cattle characterized by nodules on the skin and other parts of the body. The incubation period is 4–14 days. Epidemiological measuring of disease occurrences showed that LSD had morbidity rate can reach up to 50% while it was usually associated with low mortality rate ranged between 1 to 5%. Most of infected cattle go through febrile stage for 1-3 days during which temperature of animals reported to be (40–41.5˚C). Ocular, nasal and saliva dribbling’s from mouth commissures are variably documented (Casal et al., 2018). During LSD course, distinctive features develop due to intensive necrosis of the cutaneous nodules results in hard, raised areas known as sit-fasts. Those damaged cutaneous regions are port for entry of secondary bacterial complications causative agents and have more tendencies to myasis (Kumar et al., 2021). Secondary bacterial infection can arise, resulting in widespread suppuration and sloughing of the infected nodules. To avoid complications, antibiotics should be given to manage secondary infections, as well as adequate nursing care (Gibbs 2021). In recent years, the expansion of LSD outside of its ancestral enzootic African continent poses intense threats to the rest of the world. Quarantine measures and livestock entry regulations have been shown to be ineffective in different occasions. Vaccination with an attenuated virus is the most promising means of control, and it has been shown to be effective in preventing the spread of the disease (Abutarbush and Tuppurainen, 2018). Diseased cattle are carrying LSDV mainly in cutaneous lesions like nodules and its crusts beside its isolations form various body secretion and excretions including saliva, nasal discharge, semen, and milk (Babiuk et al., 2008). The provisional diagnosis of LSD is usually made based on the presence of specific clinical indicators, and the clinical diagnosis is validated by standard PCR (Tuppurainen et al., 2005). The genomic sequences of capripoxviruses (CaPVs) are extremely conserved, with more than 95% homology between LSD, sheep poxvirus (SPPV), and goat poxvirus (GTPV) (Kara et al., 2003). CaPVs were detected using PCR tests that used primers that targeted comparable sequences on them, with successful amplification of targets from cattle samples being classified as an LSDV and sheep or goat samples as SPP or GTPV respectively (Tuppurainen et al., 2005). Differential PCR based on primers specific to LSDV found to provide intense tool to discriminate LSDV from others members of CaPVs genus (Stram et al., 2008).
This current study aimed to investigate LSD outbreak occurred in 2019 in Sohag governorate, Egypt among Eight cattle farms of small sized herd reared under byre housing systems. Outbreak surveillance was conducted targeted cattle with apparent signs consistence LSD which were selected for sampling to confirm the disease occurrence through further viral molecular characterization. Sequencing of LSDV P32 gene segment was utilized to perform phylogenetic analysis to clarify possible LSDV strains clustering and their genetic homology. Both descriptive and molecular epidemiological tools will allow swiftly affirm the clinical diagnosis of LSD outbreaks. This will have robust impact to implement combating measures which wane the enzootic concerns of LSD in Egypt.
MATERIAL AND METHODS
Ethical Statement
The study was conducted according to guidelines of animal welfare and Research Ethics of Faculty of Veterinary Medicine; South Valley University, Egypt. During the current study, all procedures involving animals were carried out in accordance with the institutional guideline, which is based on international guidelines developed by the US National Institutes of Health (NIH publication No. 85–23, revised 1996) and the World Organization for Animal Health. Terrestrial Animal Health Code, Chapter 7.8 Use of Animals in Research, (OIE, 2015)
Rationale Of The Study
The surveillance areas were selected purposively based on the reports of LSD outbreaks occurrence between May to July 2019 at Elmonsha district, Sohag governorate, Egypt. Active outbreaks were assessed, and gathering, analysis and interpretation of surveillance, and field data were promptly documented. Clinical specimens that were collected from sick cattle were submitted to Animal Health Department, Faculty of Veterinary Medicine - South Valley University, Egypt for virological and bacteriological examinations.
Targeted Animals
The active outbreak investigation was conducted to target small dairy cattle herds having animals of different age. Cattle that showed clinical signs of Lumpy skin like lesions were targeted for this study. All animals were reared under cow byre housing and management systems. A total of 150 cow belong to 8 dairy cattle herds were examined. Detailed demographic data was provided in (Table 1).
Descriptive Epidemiological Data Of The Outbreak
Diagnosis of LSD among cattle based on the standard clinical case definition as stated elsewhere before (FAO, 2010). Infected cattle reared in Sohag Governorate, Egypt were examined clinically. These cattle suffered from fever, depression, inappetence, salivation, skin nodules and ocular-nasal discharges. Superficial lymph nodes were enlarged, especially the prescapular and precrural lymph nodes were investigated (Table 2). Morbidity and mortality was calculated.
Furthermore, all cow suspected to be infected with LDS virus and have clinical signs consistence with LSD were closely monitored and followed up for any further complication during the course of disease until recovery. The common complications were mentioned in (Table 3) in relation to treatment regimens applied and cases prognosis.
Sample Size And Sampling Collection Protocols
Along the study, a total of 150 animals were investigated. Active surveillance was conducted at the specific site of the outbreak in each dairy herd within El Monshah district. Cattle with apparent signs and suspected to be diseased with LSD were selected to be sampled.
Samples including skin biopsy; nasal swabs and saliva dribbling’s were collected from clinically affected cows and Those samples are used for molecular detection of LSD Virus and identification of the bacterial species incorporated to cause secondary bacterial infection among cattle. In addition, bedding/manure samples were collected from the floor of the cow byre house for isolation and identification of the environmental bacterial agents causing secondary complications.
All samples were collected aseptically as described before (OIE, 2018). Skin nodules from cattle which showed clinical signs of the disease were taken aseptically after washing and cleaning the area and removing the hairs with the help of sterile scalpel blade. Samples of skin biopsies and scabs were collected in sterile cryovials containing Minimum Essential Medium (MEM, Merck-Sigma, USA). The nasal swabs were taken aseptically, placed in the sterilized cryovial contain MEM. Saliva dribbling was aseptically collected in sterile vials containing MEM. Each sample was given a unique sample ID, placed in an ice cooler box and transferred to Animal Health Department, Faculty of Veterinary Medicine, South Valley University to be stored at − 80 °C for further molecular analysis.
Bedding Samples
A total of twenty bedding samples were collected according to Rendos et al. (1975). The total amount of material collected in sterile plastic bags. Samples for microbiological analyses were prepared by adding 10 g of the thorough
Table 1: Demographic data of the affected herds (N=150 cow).
| Location | Housing system | Species | Age (years) | Gender | Herd size | Problem of Ticks | Vaccination |
| 1 | Cow byre system | Dairy cow | 1-3 | Female | 15 | none | unvaccinated |
|
2 |
Cow byre system | Dairy cow | 2-4 | Female | 19 | none | unvaccinated |
|
3 |
Cow byre system | Dairy cow | 1-4 | Female | 18 | none | vaccinated |
|
4 |
Cow byre system | Dairy cow | 2–3 | Female | 24 | yes | unvaccinated |
|
5 |
Cow byre system | Dairy cow | 1–4 | Female | 22 | none | unvaccinated |
|
6 |
Cow byre system | Dairy cow | 1–3 | Female | 21 | yes | vaccinated |
|
7 |
Cow byre system | Dairy cow | 2–4 | Female | 16 | none | unvaccinated |
|
8 |
Cow byre system | Dairy cow | 3–4 | Female | 15 | yes |
vaccinated |
Table 2: Descriptive Epidemiology of outbreak among affected herds
| Location | Number of clinically infected cattle | Morbidity | Mortality | Predominate Clinical Signs | Types Of Specimens Collected |
| 1 | 4 | 26.6% | 0% | Pyrexia, Cutaneous nodules, lymph node enlargements | Skin nodules biopsy, Nasal swabs , bedding material |
|
2 |
6 | 31.5% | 5% | Pyrexia, Lacrimation, Cutaneous nodules, salivation | Skin nodules biopsy, Nasal swabs , bedding material |
|
3 |
3 | 16.5% | 0% |
Pyrexia, Nasal discharge, Cutaneous nodules |
Skin nodules biopsy, Nasal swabs , bedding material |
|
4 |
5 | 20.8% | 0% |
Pyrexia, saliva dribbling, Cutaneous nodules |
Skin nodules biopsy, Nasal swabs , bedding material |
|
5 |
7 | 31.8% | 4% |
Pyrexia, respiratory distress, Cutaneous nodules, salivation |
Skin nodules biopsy, Nasal swabs , bedding |
|
6 |
4 | 19% | 0% | Pyrexia, Cutaneous nodules, lymph node enlargements | Skin nodules biopsy, Nasal swabs , bedding material |
|
7 |
6 | 37.5% | 0% |
Pyrexia, saliva dribbling , Cutaneous nodules |
Skin nodules biopsy, Nasal swabs , bedding material |
|
8 |
3 | 20% | 0% |
Pyrexia, saliva dribbling, Cutaneous nodules, lymph node enlargements |
Skin nodules biopsy, Nasal swabs , bedding material |
Table 3: Types and percentage of complications among 38 animals suffered from LSD during outbreak
| Complications | Prevalence | Treatment options used | Prognosis |
| Lameness | 18% | Long acting antibiotics + anti-inflammatory | good |
|
Recumbency |
2.6% | Supportive therapy and tonics | poor |
|
Eye affection |
13% | Topical ointments/drops | good |
|
Pneumonia |
15.7% | Long acting antibiotics + anti-inflammatory | good |
|
Abortion |
0% | ----- | ----- |
|
Mastitis |
21% | Udder and intramammary infusion antibiotics | good |
|
Edema of limbs or other body parts |
26% | Long acting antibiotics + anti-inflammatory | good |
|
Complicated Necrotic nodules/wounds |
13% | Long acting antibiotics |
good |
| 15.7% | Long acting antibiotics + topical antiseptics + dressing |
Very good |
ly mixed wet bedding sample to 90 ml of sterile PBS in sterile glass bottle. Bottles were then mixed thoroughly by manual agitation by swinging the bottles from side to side for 5 min. After the mixture stored for 30 min, the supernatant was collected and centrifuged at 5,000×g for 5 min at room temperature. The pellet was resuspended with 1 mL of PBS.
Dna Extraction and Pcr Confirmation of Lsdv
At room temperature, skin biopsies, nasal and saliva samples were thawed. Skin biopsy samples were cut into tiny pieces with a sterile scalpel blade, weighed, and homogenised in 500 liter of sterile 1X PBS solution, pH 7.4. Total DNA was extracted from tissue homogenates, nasal and saliva samples using a DNeasy Blood and Tissue kit (Qiagen, Germany) according to the manufacturer’s instructions. Following that, PCR was performed; forward primer, 5′-TTTCCTGATTTTTCTTACTAT-3′ and reverse primer, 5′-AAATTATATACGTAAATAAC-3′, and PCR conditions as described by Ireland and Binepal (1998). PCR amplification was done in a standard 25 μl PCR reaction. The reaction mixture included 12.5 μl of 2× Taq universal master mix (Thermo Fisher Scientific, USA), 6.5 μl deionized water, 0.5 μl of P32 gene forward primer and 0.5 μl of P32 gene reverse primer and 5 μl of the DNA template. The primers were developed from the viral attachment protein encoding gene according to (Ireland and Binepal 1998).
Sequencing Of Amplified Pcr Products
Purified PCR products were sequenced using an automated ABI3500XL Genetic Analyzer and BigDye terminator v3.1 cycle sequencing reactions (Applied Biosystems, Foster City, CA), according to the manufacturer’s instructions at the Animal Health Department, Faculty of Veterinary Medicine, South Valley University. The same primers used to carry out the amplification of the P32 gene in the lab were also used in the sequencing reactions. The sequence analysis and alignments of P32 gene of LSDV1-14/sohag/Egypt 2019 was done using BioEdit software version 7. For evolutionary analysis, corresponding sequences of other LSDV strains, SPV and GPV strains were retrieved from GenBank. Phylogenetic analyses were carried out using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates). The evolutionary distances were computed using the Maximum Composite Likelihood method (Kumar et al., 2018).
The analysis involved a total of 29 nucleotide sequences for P32/LSDV including 14 LSDV from the current study and 8 P32 LSDV sequences obtained from GenBank: MN598005.1, MN422451.1, MN422450.1, MN422449.1, MN422448.1, MN422447.1, KX960769.1 and KX960782.1. Goat Pox References strains AY382869.1, KY508697.1, MK948083.1 and MG458382.1 and Sheep pox SPV including KT964233.1, MG458370.1 and KC847056.1 were retrieved form GenBank and used for rooting the phylogenetic relationships among strains.
Isolation and Identification of Pathogenic Enterobacteriaceae
All samples were inculcated into MacConkey broth for enrichment. The inoculated samples were incubated at 37º C for 24hours. A loopful was streaked on Eosin Methylene blue agar and MacConkey agar. The inoculated plates were incubated at 37º C for 24-48 hours. Furthermore, after enrichment, each sample was inculcated on two sets of Violet Red Bile (VRB) agar plates which then were incubated at 37 ºC and 44.5 ºC for 24 h for differentiation of pathogenic fecal coliform from others nonpathogenic. Suspected growing colonies on MacConkey agar, Eosin Methylene blue agar medium and VRB agar were carefully picked-up and slanted on nutrient agar slopes for further biochemical identification tests.
Isolation And Identification Of Staphylococcus Species
The International Organization for Standardization approved techniques were used for Staphylococci and S. aureus isolation and identification (ISO: 6888-1).
Isolation of total Staphylococci and S. aureus were performed using both Manitol Salt Agar and Baird-Parker (BP) agar medium (CM 275, Oxoid, UK) supplemented with egg yolk tellurite emulsion (SR 54, Oxoid, UK). The plates were inoculated with 0.1 mL of original prepared samples using plating technique. Inoculated plates were incubated at 37°C for 24-48 h. The colonies were tentatively recognized as Coagulase-positive Staphylococcus on the basis of a shift in pH in the medium (red to yellow color) on for Manitol Salt Agar while on Baird-Parker (BP) agar, the usual black colonies surrounded by an opaque halo are thought to be presumptive S. aureus. The representative colonies were subculture on nutrients slants and incubated at 37 C. The slant was preserved and maintained for further characterizing the isolates.
RESULTS
Demographic Data Analysis
The study is a part of active epidemiological surveillance initiated during May to July 2019 as response to LSD outbreak among cattle reared in small herds using cow byre system. All animals are tied up in their stalls on dirty floor along the day in tail to tail arrangements with manger in front of each animal for rations while water presented to the animals in buckets periodically. The herds sizes are ranged from 15 to 24 animals per herd with age ranged from 1 to 4 years. Vaccination state against LSDV are variable, some herds are vaccinated (37.5%) while others didn’t receive any vaccination (62.5%) during the last one year before outbreak (Table 1).
DESCRIPTIVE EPIDEMIOLOGY OF THE OUTBREAK
Measuring of outbreak occurrence and clinical findings: The overall prevalence of lumpy skin virus infection between May to July 2019 among all the 8 investigated herds was 38/150 (25.3%). In addition, the prevalence in-between herds were ranged from 16.5% to 37.5%. Mortality was low among infected cattle and ranged from 4 to 5% and reported only in two herds (Table 2).
During the initial phase of the diseases, most of animal suffered from fever with animal body temperature ranged to be (40.5 to 41°C). Along the febrile stages which in majority of cases persisted for 3 to 4 days, cows had Anorexia and some infected cattle (44%) exhibits salivation, lacrimation (15%) and nasal discharge (7%). The most prominent clinical finding was the appearance of cutaneous nodules affecting either certain parts or /all over the body in 75% infected animals (Figure 1). The nodules are well circumscribed, round, slightly raised, firm, and painful. Some nodules were developing on the muzzle and within the nasal and buccal mucous membranes. The skin nodules contain a firm, creamy-gray or yellow mass of tissue. Clinical examination of animals revealed that (26%) had enlarged regional lymph nodes somewhere in their body; mostly the prescapular and prefemoral lymph nodes were involved.
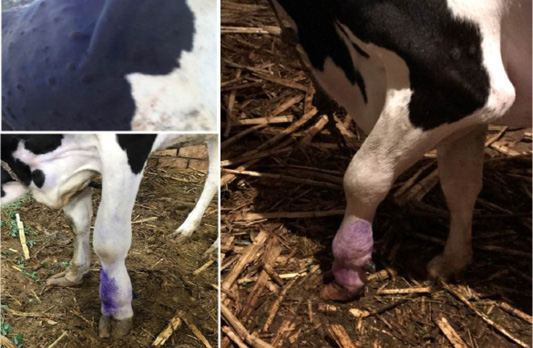
Figure 1: Lumpy skin disease in cow showing (a): Cutaneous nodules (b): marked edema of left forelimb and Enlargement of prescapular lymph node (c): edema of brisket and sever lameness
Complications of lsd among infected cows during outbreak: Most of the diseased cattle were recovered except few which died during the infectious period of the disease. Oedema of fore or hind limbs and other parts of the body including brisket were observed in 26% of cases accompanied with various degree of lameness in 18% of the affected cows (Figure 1). Some of cattle suffered from lameness have incoordination gate and higher tendency for recumbency. Using the cow byre system with dirty floor was reasonable factor make secondary bacterial infection more liable to increase the incidence of mastitis and cutaneous nodules complications. Some of the cutaneous nodules got cracked, ruptured and create a deep-seated wound. Wounds were frequently invaded by secondary bacterial infections which led to extensive suppuration and sloughing. The lesions were particularly extensive in the foreshank/ fetlock regions, extended up to the underlying subcutis and muscle (Figure 2). Others types and percentage of complications among cattle suffered from LSD during investigated outbreak was listed in (Table 3).
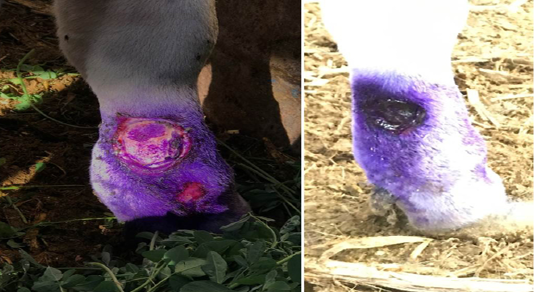
Figure 2: Cattle infected with LSD showing (a) open, ulcerated and bleeding cutaneous lesions filled with pus in the lower parts of the forelimb. The ruptured nodules created a deep-seated wound; invaded by secondary bacterial infection leading to suppuration and sloughing, lesions extending to the underlying subcuitis and muscle (b) after staring antibiotic treatment, topical antiseptics and signs of healing is clearly noticed.
Molecular Identification of the Lsdv Using Pcr Amplification of P32 Gene
The P32 gene was amplified using conventional gel-based PCR. In positive samples, the amplicon size of the PCR product had a molecular weight of 192 bp for the attachment protein gene, which was equal to the expected amplification product size from the reference LSDV (Figure 3). The P32 gene was successfully amplified in 60/95 (63%) of all examined samples. LSDV was detected in 73, 63, and 52 samples from skin nodules, saliva dribbling and nasal discharge, respectively (Table 4).
Table 4: Detection of lumpy skin virus in skin nodules biopsy, saliva and nasal discharge using PCR
| Specimens | Total samples | Incidences in various samples | |
| Number |
%
|
||
| Skin nodules biopsy | 38 | 28 |
73 |
| Nasal swabs/discharge | 38 | 20 |
52 |
| Saliva droppings | 19 | 12 |
63 |
Lsdv Multiple Sequence Alignments And Phylogenetic Analysis
Multiple sequence alignments of the partial nucleotide sequences of P32 gene were done using BioEdit software Version7 (Figure 4). Phylogenetic reconstruction of the
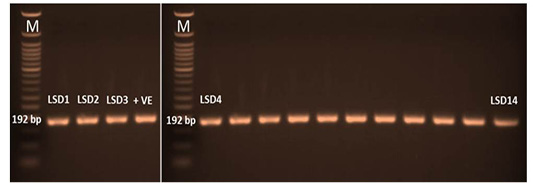
Figure 3: PCR amplicons (192 bp) of P32 gene LSDV. LSD1 till lane 14 were the amplified samples. Lane M was 50 bp DNA ladder and lane +v represent positive control
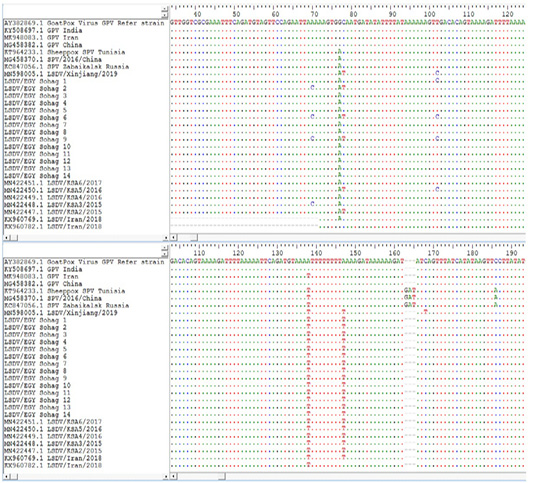
Figure 4: Multiple sequence alignments of the partial nucleotide sequences of P32 gene were done using BioEdit software Version7. LSDVs from Sohag, Egypt were aligned with Goat pox reference strains and representative LSDVs’ sequences retrieved from GenBank. Identical nucleotides are indicated with dots.
P32 gene sequences was done using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates). The evolutionary distances were computed using the Maximum Composite Likelihood method. (Figure 5). The derived phylogenetic tree had four main clusters. Cluster I had most of the Sohag Egyptian isolates including strains LSDV/EGY Sohag 3,4,5,7,8,10,11. Cluster II composed of two strains LSDV/EGY Sohag 6, 9, 12, 2. Cluster III clarified the similarity between LSDV/EGY Sohag 1, 13, 14. Phylogenetically, some strain share high degree of homology with some others Egyptian strains and Saudi Arabia strains.
Prevalence of Bacterial Species Often Complicate Burst Necrotic Nodules Among Infected Lsd Cattle
Isolation of bacterial species suspected to cause secondary
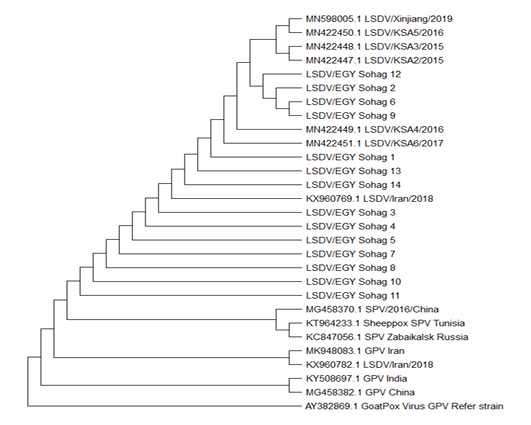
Figure 5: The sequence analysis and alignments of P32 gene of LSDV1-14/sohag/Egypt 2019 was done using BioEdit software version 7. For evolutionary analysis, corresponding sequences of other LSDV strains, SPV and GPV strains were retrieved from GenBank. Phylogenetic analyses were carried out using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates). The evolutionary distances were computed using the Maximum Composite Likelihood method.
Table 5: Prevalence of bacterial species often complicate burst necrotic nodules among infected LSD cattle.
| Bacterial species causing secondary infection | Prevalence in various samples | |||
| Bedding n=38 | Nasal swab n=38 | Necrotic nodules n=38 | ||
| Staphylococci species | Overall species | 20% | 40% |
36% |
|
Staph. aureus |
11% | 26% | 31% | |
|
Fecal Coliform |
Overall species | 80% | 10% |
60% |
|
E. coli species |
65% | 6% |
45% |
|
|
Klebsiella species |
35% | 4% |
38% |
|
infection was performed to throw a beam of light about their role in prognosis and complete recovery of infected animal. Staphylococci species were isolated from 36% of animals that have skin nodules complication manifested in the form of supportive discharge and sometimes destructive necrotic lesion, tissue sloughing and massive pus discharge. S. aureus was isolated at prevalent of 26% form nodules of animals that exhibit some kind of complication. In addition, about 40% of examined animals found to harbor Staphylococci species in their nasal discharge. Furthermore, checking the role of environmental fecal coliform and their liability to cause secondary bacterial infection in infection cattle was clarified. The results highlighted that fecal coliform were isolated from complicated nodules among 60% of the infected cattle. E. coli species could be recovered form infected skin nodules in 45% of infected animals (Table 5).
DISCUSSION
Devastating consequences on health and production of cattle were reported due to viral diseases like LSD which reported to be enzootic over decades in African continent. The disease was recently jumped out Africa to involve numerous countries in the Middle East like Egypt (Tuppurainen et al., 2015). In Egypt, cattle industry is negatively impacted due to affection with enzootic viral infectious diseases like LSD (Ahmed et al., 2021). Egyptian contingency plan for combating of LSD is depending on mass vaccination of susceptible host with sheep pox vaccine manufactured at the Veterinary Serum and Vaccine Research Institute, Egypt. Despite the huge efforts to control the re-emergency of LSD in Egypt, the susceptible hosts are challenged with outbreaks of various temporal and spatial patterns resulting in devastating consequences on livestock industry (Allam et al., 2020). Chronological trace back of the LSD epidemiology in Egypt along the last decade revealed that destructive LSD outbreak has occurred among cattle in different localities of Egypt during 2006 (El-Kholy et al., 2008). Since that, multiple outbreaks have reemerged in different Egyptian governorates. However the disease was reported in Egypt since 1988, during which there were two outbreaks had been spread in eastern region of Egypt (House et al., 1990). Taking earlier reports and the finding of LSD cases in cattle in this research into account, it is apparent that this economically disastrous epidemic is expanding its geographic cover spread in Egypt and presents a risk to Eastern Europe, Russia and Asia. (Elhaig et al., 2017).The frequent occurrence of LSD outbreaks emphasize the vital role of ongoing active surveillance programs which is key component incorporated at every combating program to measure the disease occurrence. To understand how such a virus has been adapted to be endemic in Egypt still need an extensive characterization of LSDV locally and in neighboring countries which could help to resolve the re-emergence of these isolates (Figure 6).
During the current study, cattle belong to eight small sized herds were investigated during the LSD outbreak, 2019. Cattle were aged between 1 to 4 years which reared under poor husbandry practices, tied up in their stalls on dirty floor along the day in tail to tail arrangements. Lack of measures for insect vectors control were common among most of herds. Haematophagous insects, such as mosquitoes and stable flies were well documented that have main role in transmission of the LSDV (Lubinga et al., 2014). Ixodid ticks have recently been suspected of playing a role in the transmission of LSDV in cattle (Tuppurainen et al., 2011). Milking practice and others managemental procedures that result in direct contact between cattle were reported to have a role in spreading LSDV (Magori-Cohen et al. 2012).

Figure 6: Spatial and temporal lumpy skin diseases occurrences patterns In Egypt, Africa and globally. (A) Left figure represent the outbreak of LSD 2019 in Egypt that investigated during the current study in relation to others outbreaks occurred at African continent. (B) Maps showing chronological epizootic events in Africa and the transboundary potential spreads of LSD globally {Upper right maps, 2006 and lower right map, 2015}. Data was extracted form (OIE-WAHIS, 2021)
Vaccination rate among herds are relatively low (37.5%) while majority of herds representing 62.5% didn’t receive any vaccines against LSV. Those factors including poor insect vector control strategies and uneven vaccination coverage might contribute largely to the reoccurrences episodes of LSD among Egyptian cattle. This was in agreement with recent study stated that, the current vaccine strategy in Egypt should be re-evaluated for more coverage and effectiveness (Ahmed et al., 2021).
The diagnosis of LSD was based on clinical findings, confirmed by PCR and the viruses in samples collected from outbreaks between May to July 2019 in Egypt were phylogenetically tracked. All the investigated herds were affected during the outbreak and showed variable prevalence among herds ranged from 16.5% to 37.5%. This was consistence with former reports clarified; the intra-herd prevalence rate of clinical cases was 17.3% and mostly in adult animals (Elhaig et al., 2017). Majority of the affected cattle exhibited common clinical manifestations of LSD in cattle. This was agreed well with other studies found that suspected clinical signs were consistent with LSDV infection (Elhaig et al., 2017). High grade fever that might reach 41°C were predominate among most infected cattle could last for few during which animals’ suffered from anorexic condition. During the febrile stage, loss of appetite and decreasing the feed intake greatly affected the health conditions of the infected cows. Skin lesions including the characteristics lumps and nodules were developed in 75% of the affected cattle. Most of those skin nodules matches the common features that consistence with LSD. This was harmonizing with previous reports mentioned that All suspected cattle exhibited cutaneous lesions including nodules and sitfast which were significantly suggestive clinical findings for LSD (Ahmed et al., 2021). Salivation was documented in (44%) of the examined animals while 15% and 7% of the infected cattle presented suffering from marked lacrimation and nasal discharge, respectively. Based on clinical examination, lymphadenopathy mostly involved the prescapular and prefemoral lymph nodes enlargements were noticed in 26% of infected animals. Hence, descriptive epidemiology of LSD epidemic attacks based on measuring of outbreak occurrence and clinical findings still represents a corner stone of filed LSD diagnosis required for rapid implementations of appropriate control regimen and avoid dissemination of LSDV strains among the Egyptian susceptible livestock. The presence of LSD in cattle necessitates more epidemiological research on the disease’s transmission in the area, as well as the implementation of control and preventative tactics (Elhaig et al., 2017).
Assuring the clinical suspicious was done using a conventional PCR targeting to amplify 192 bp of P32 gene in LSDV. In the present study, skin nodules specimens out of examined animals prove that 73% were positive for LSDV. This was in agreement with former study revealed that cutaneous nodules and other skin lesions tend to have higher LSD viral load and can persist for long period so it was more superior to any others organs for the detection of viral DNA of LSDV (Zeynalova et al., 2016). Historically, Egypt has used to import cattle from various origin including Africa and globally. Hence, trace back the origin of the LSD virus through sequence analysis of conserved genetic loci is common epidemiological tool to can track the transboundary potential of the diseases (Sharawi and Abd El-Rahim, 2011).
The phylogenetic tree based on P32 gene sequences resulted in drawing of several clusters among the isolates of the current study (Figure 5). Overall, most of isolates exhibit strong degree of phylogenetic relatedness indicating that strains of LSDV circulating in Egypt, has well established enzootic state. Many strains LSDV/EGY Sohag like 12, 2, 6 and 9 are found to have close phylogenetic correlation and share homology with strains form Saudi Arabia.
In Egypt, cattle infected with LSDV receive treatment symptomatically according the stage of the disease and severity of the case. As common practice, diseased cow during the initial febrile stage are treated with antipyretics and anti-inflammatory, closely monitored for any relevant other supportive treatments. Upon appearances of any complication, especially secondary bacterial infection, administration of broad spectrum antibiotics is initiated. The predominant bacterial species incorporated in secondary complications of cutaneous lesion among LSD infected animals were Staphylococci species (36%), Staph. aureus (26 %), fecal coliform (60%) and E. coli species (45 %). In our study, we found that some cattle developed sever lameness due to extensive suppuration and sloughing of the cutaneous nodules at fore/hind limbs. Those developed such conditions were 11 cows divided into two groups. The First group comprised of Five cow that were treated with long acting antibiotics for 5-7 days administrated intramuscularly with frequent drainage of the pus from the infected purulent cutaneous nodules. The prognosis was good however, those group taken longer time for recovery and healing process. The second group had six cows that therapeutically managed with some modification. All cows were injected with broad spectrum long acting antibiotics and the purulent cutaneous nodules were managed after pus drainage with topical application of antiseptics two times per day 5-7 days. The prognosis was very good and the recovery, healing process took shorted time than the first group.
CONCLUSIONS
LSD has enzootic potential in Egypt. Multi-targeted approach, including descriptive and molecular epidemiological tools allows accurate and rapid techniques to affirm the clinical diagnosis of LSD outbreaks. This will help in the early containment of an outbreak; facilitate rapid application of combating measures in enzootic area to preventing the dissemination of the disease during epizootic attacks. This work emphasizes the need of continued surveillance and tracing of the circulating LSDV strains, as well as the necessity to constantly modify local and worldwide contingency plans according to the disease pattern and the factors affecting its occurrences.
ACKNOWLEDGMENTS
The authors would like to acknowledge Department of Animal Heath, Faculty of veterinary medicine, South Valley University. Authors extend their appreciation to farms owners: Abd El-Rahman Abdellatef Diab and others for their valuable cooperation.
CONFLICT OF INTEREST
All the authors declare that they have no conflict of interest.
AUTHORS’ CONTRIBUTION
HMD designed the study and edited the manuscript. HMA and ASA collected the samples. All authors shared in the lab work, performed the interpretation of the results. The final version of the manuscript was read and approved by all authors.
REFERENCES





